Rethinking how DNA methylation patterns are maintained
- PMID: 19789556
- PMCID: PMC2848124
- DOI: 10.1038/nrg2651
Rethinking how DNA methylation patterns are maintained
Abstract
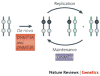
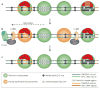
DNA methylation patterns are set up early in mammalian development and are then copied during the division of somatic cells. A long-established model for the maintenance of these patterns explains some, but not all, of the data that are now available. We propose a new model that suggests that the maintenance of DNA methylation relies not only on the recognition of hemimethylated DNA by DNA methyltransferase 1 (DNMT1) but also on the localization of the DNMT3A and DNMT3B enzymes to specific chromatin regions that contain methylated DNA.
Figures
References
-
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. - PubMed
-
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. - PubMed
-
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. - PubMed
-
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. - PubMed
-
- Smith HO, Kelly SV. Methylases of the Type II Restriction-Modification Systems. In: Razin A, Cedar H, Riggs AD, editors. DNA Methylation Biochemistry and Biological Significance. Springer-Verlag; New York: 1984. pp. 39–71.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

