Molecular risk stratification in advanced heart failure patients
- PMID: 19793385
- PMCID: PMC3829011
- DOI: 10.1111/j.1582-4934.2009.00913.x
Molecular risk stratification in advanced heart failure patients
Abstract
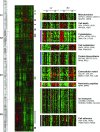
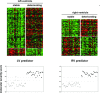
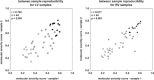
Risk stratification in advanced heart failure (HF) is crucial for the individualization of therapeutic strategy, in particular for heart transplantation and ventricular assist device implantation. We tested the hypothesis that cardiac gene expression profiling can distinguish between HF patients with different disease severity. We obtained tissue samples from both left (LV) and right (RV) ventricle of explanted hearts of 44 patients undergoing cardiac transplantation or ventricular assist device placement. Gene expression profiles were obtained using an in-house microarray containing 4217 muscular organ-relevant genes. Based on their clinical status, patients were classified into three HF-severity groups: deteriorating (n= 12), intermediate (n= 19) and stable (n= 13). Two-class statistical analysis of gene expression profiles of deteriorating and stable patients identified a 170-gene and a 129-gene predictor for LV and RV samples, respectively. The LV molecular predictor identified patients with stable and deteriorating status with a sensitivity of 88% and 92%, and a specificity of 100% and 96%, respectively. The RV molecular predictor identified patients with stable and deteriorating status with a sensitivity of 100% and 96%, and a specificity of 100% and 100%, respectively. The molecular prediction was reproducible across biological replicates in LV and RV samples. Gene expression profiling has the potential to reproducibly detect HF patients with highest HF severity with high sensitivity and specificity. In addition, not only LV but also RV samples could be used for molecular risk stratification with similar predictive power.
Figures

References
-
- Lee DS, Austin PC, Rouleau JL, et al. Predicting mortality among patients hospitalized for heart failure: derivation and validation of a clinical model. JAMA. 2003;290:2581–7. - PubMed
-
- Levy WC, Mozaffarian D, Linker DT, et al. The Seattle Heart Failure Model: prediction of survival in heart failure. Circulation. 2006;113:1424–33. - PubMed
-
- Aaronson KD, Schwartz JS, Chen TM, et al. Development and prospective validation of a clinical index to predict survival in ambulatory patients referred for cardiac transplant evaluation. Circulation. 1997;95:2660–7. - PubMed
-
- Smits JM, Deng MC, Hummel M, et al. A prognostic model for predicting waiting-list mortality for a total national cohort of adult heart-transplant candidates. Transplantation. 2003;76:1185–9. - PubMed
-
- Francis GS. Pathophysiology of chronic heart failure. Am J Med. 2001;110:37S–46S. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

