The cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein gp41 harbors lipid raft association determinants
- PMID: 19793805
- PMCID: PMC2798425
- DOI: 10.1128/JVI.00899-09
The cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein gp41 harbors lipid raft association determinants
Abstract
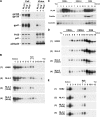
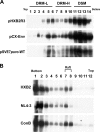
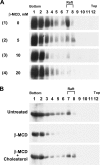
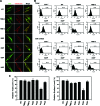
The molecular basis for localization of the human immunodeficiency virus type 1 envelope glycoprotein (Env) in detergent-resistant membranes (DRMs), also called lipid rafts, still remains unclear. The C-terminal cytoplasmic tail of gp41 contains three membrane-interacting, amphipathic alpha-helical sequences, termed lentivirus lytic peptide 2 (LLP-2), LLP-3, and LLP-1, in that order. Here we identify determinants in the cytoplasmic tail which are crucial for Env's association with Triton X-100-resistant rafts. Truncations of LLP-1 greatly reduced Env localization in lipid rafts, and the property of Gag-independent gp41 localization in rafts was conserved among different strains. Analyses of mutants containing single deletions or substitutions in LLP-1 showed that the alpha-helical structure of the LLP-1 hydrophobic face has a more-critical role in Env-raft associations than that of the hydrophilic face. With the exception of a Pro substitution for Val-833, all Pro substitution and charge-inverting mutants showed wild-type virus-like one-cycle viral infectivity, replication kinetics, and Env incorporation into the virus. The intracellular localization and cell surface expression of mutants not localized in lipid rafts, such as the TM844, TM813, 829P, and 843P mutants, were apparently normal compared to those of wild-type Env. Cytoplasmic subdomain targeting analyses revealed that the sequence spanning LLP-3 and LLP-1 could target a cytoplasmic reporter protein to DRMs. Mutations of LLP-1 that affected Env association with lipid rafts also disrupted the DRM-targeting ability of the LLP-3/LLP-1 sequence. Our results clearly demonstrate that LLP motifs located in the C-terminal cytoplasmic tail of gp41 harbor Triton X-100-resistant raft association determinants.
Figures





Similar articles
-
Cellular membrane-binding ability of the C-terminal cytoplasmic domain of human immunodeficiency virus type 1 envelope transmembrane protein gp41.J Virol. 2001 Oct;75(20):9925-38. doi: 10.1128/JVI.75.20.9925-9938.2001. J Virol. 2001. PMID: 11559825 Free PMC article.
-
Rational site-directed mutations of the LLP-1 and LLP-2 lentivirus lytic peptide domains in the intracytoplasmic tail of human immunodeficiency virus type 1 gp41 indicate common functions in cell-cell fusion but distinct roles in virion envelope incorporation.J Virol. 2003 Mar;77(6):3634-46. doi: 10.1128/jvi.77.6.3634-3646.2003. J Virol. 2003. PMID: 12610139 Free PMC article.
-
HIV-1 Cell-Free and Cell-to-Cell Infections Are Differentially Regulated by Distinct Determinants in the Env gp41 Cytoplasmic Tail.J Virol. 2015 Sep;89(18):9324-37. doi: 10.1128/JVI.00655-15. Epub 2015 Jul 1. J Virol. 2015. PMID: 26136566 Free PMC article.
-
The frantic play of the concealed HIV envelope cytoplasmic tail.Retrovirology. 2013 May 24;10:54. doi: 10.1186/1742-4690-10-54. Retrovirology. 2013. PMID: 23705972 Free PMC article. Review.
-
HIV-1 replication.Somat Cell Mol Genet. 2001 Nov;26(1-6):13-33. doi: 10.1023/a:1021070512287. Somat Cell Mol Genet. 2001. PMID: 12465460 Review.
Cited by
-
Analysis of herpes simplex type 1 gB, gD, and gH/gL on production of infectious HIV-1: HSV-1 gD restricts HIV-1 by exclusion of HIV-1 Env from maturing viral particles.Retrovirology. 2019 Apr 2;16(1):9. doi: 10.1186/s12977-019-0470-5. Retrovirology. 2019. PMID: 30940160 Free PMC article.
-
Localization to detergent-resistant membranes and HIV-1 core entry inhibition correlate with HIV-1 restriction by SERINC5.Virology. 2018 Feb;515:52-65. doi: 10.1016/j.virol.2017.12.005. Epub 2017 Dec 18. Virology. 2018. PMID: 29268082 Free PMC article.
-
Cholesterol in the Viral Membrane is a Molecular Switch Governing HIV-1 Env Clustering.Adv Sci (Weinh). 2020 Dec 21;8(3):2003468. doi: 10.1002/advs.202003468. eCollection 2021 Feb. Adv Sci (Weinh). 2020. PMID: 33552873 Free PMC article.
-
Topology of the C-terminal tail of HIV-1 gp41: differential exposure of the Kennedy epitope on cell and viral membranes.PLoS One. 2010 Dec 7;5(12):e15261. doi: 10.1371/journal.pone.0015261. PLoS One. 2010. PMID: 21151874 Free PMC article.
-
The role of matrix in HIV-1 envelope glycoprotein incorporation.Trends Microbiol. 2014 Jul;22(7):372-8. doi: 10.1016/j.tim.2014.04.012. Epub 2014 Jun 2. Trends Microbiol. 2014. PMID: 24933691 Free PMC article. Review.
References
-
- Anderson, R. G. 1998. The caveolae membrane system. Annu. Rev. Biochem. 67:199-225. - PubMed
-
- Arreaza, G., and D. A. Brown. 1995. Sorting and intracellular trafficking of a glycosylphosphatidylinositol-anchored protein and two hybrid transmembrane proteins with the same ectodomain in Madin-Darby canine kidney epithelial cells. J. Biol. Chem. 270:23641-23647. - PubMed
-
- Berlioz-Torrent, C., B. L. Shacklett, L. Erdtmann, L. Delamarre, I. Bouchaert, P. Sonigo, M. C. Dokhelar, and R. Benarous. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73:1350-1361. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

