Populations of genomic RNAs devoted to the replication or spread of a bipartite plant virus differ in genetic structure
- PMID: 19793810
- PMCID: PMC2786829
- DOI: 10.1128/JVI.00950-09
Populations of genomic RNAs devoted to the replication or spread of a bipartite plant virus differ in genetic structure
Abstract
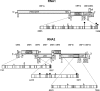
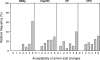
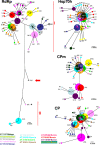
RNA viruses within a host exist as dynamic distributions of closely related mutants and recombinant genomes. These closely related mutants and recombinant genomes, which are subjected to a continuous process of genetic variation, competition, and selection, act as a unit of selection, termed viral quasispecies. Characterization of mutant spectra within hosts is essential for understanding viral evolution and pathogenesis resulting from the cooperative behavior of viral mutants within viral quasispecies. Furthermore, a detailed analysis of viral variability within hosts is needed to design control strategies, because viral quasispecies are reservoirs of viral variants that potentially can emerge with increased virulence or altered tropism. In this work, we report a detailed analysis of within-host viral populations in 13 field isolates of the bipartite Tomato chlorosis virus (ToCV) (genus Crinivirus, family Closteroviridae). The intraisolate genetic structure was analyzed based on sequencing data for 755 molecular clones distributed in four genomic regions within the RNA-dependent RNA polymerase (RNA1) and Hsp70h, CP, and CPm (RNA2) open reading frames. Our results showed that populations of ToCV within a host plant have a heterogeneous and complex genetic structure similar to that described for animal and plant RNA viral quasispecies. Moreover, the structures of these populations clearly differ depending on the RNA segment considered, being more complex for RNA1 (encoding replication-associated proteins) than for RNA2 (encoding encapsidation-, systemic-movement-, and insect transmission-relevant proteins). These results support the idea that, in multicomponent RNA viruses, function can generate profound differences in the genetic structures of the different genomic segments.
Figures
Similar articles
-
Tomato chlorosis virus, an emergent plant virus still expanding its geographical and host ranges.Mol Plant Pathol. 2019 Sep;20(9):1307-1320. doi: 10.1111/mpp.12847. Epub 2019 Jul 2. Mol Plant Pathol. 2019. PMID: 31267719 Free PMC article.
-
Complete nucleotide sequence of the RNA2 of the crinivirus tomato chlorosis virus.Arch Virol. 2006 Mar;151(3):581-7. doi: 10.1007/s00705-005-0690-y. Epub 2005 Dec 28. Arch Virol. 2006. PMID: 16374719
-
Multiple suppressors of RNA silencing encoded by both genomic RNAs of the crinivirus, Tomato chlorosis virus.Virology. 2008 Sep 15;379(1):168-74. doi: 10.1016/j.virol.2008.06.020. Epub 2008 Jul 21. Virology. 2008. PMID: 18644612
-
RNA virus quasispecies: significance for viral disease and epidemiology.Infect Agents Dis. 1994 Aug;3(4):201-14. Infect Agents Dis. 1994. PMID: 7827789 Review.
-
Historical Perspective on the Discovery of the Quasispecies Concept.Annu Rev Virol. 2021 Sep 29;8(1):51-72. doi: 10.1146/annurev-virology-091919-105900. Annu Rev Virol. 2021. PMID: 34586874 Review.
Cited by
-
High variability and rapid evolution of a nanovirus.J Virol. 2010 Sep;84(18):9105-17. doi: 10.1128/JVI.00607-10. Epub 2010 Jun 30. J Virol. 2010. PMID: 20592071 Free PMC article.
-
Genetic background matters: a plant-virus gene-for-gene interaction is strongly influenced by genetic contexts.Mol Plant Pathol. 2011 Dec;12(9):911-20. doi: 10.1111/j.1364-3703.2011.00724.x. Epub 2011 Jun 1. Mol Plant Pathol. 2011. PMID: 21726391 Free PMC article.
-
Genetic diversity and silencing suppression activity of the p22 protein of Tomato chlorosis virus isolates from tomato and sweet pepper.Virus Genes. 2015 Oct;51(2):283-9. doi: 10.1007/s11262-015-1244-3. Epub 2015 Sep 3. Virus Genes. 2015. PMID: 26334965
-
Genetic variability and evolutionary dynamics of viruses of the family Closteroviridae.Front Microbiol. 2013 Jun 26;4:151. doi: 10.3389/fmicb.2013.00151. eCollection 2013. Front Microbiol. 2013. PMID: 23805130 Free PMC article.
-
Tomato chlorosis virus, an emergent plant virus still expanding its geographical and host ranges.Mol Plant Pathol. 2019 Sep;20(9):1307-1320. doi: 10.1111/mpp.12847. Epub 2019 Jul 2. Mol Plant Pathol. 2019. PMID: 31267719 Free PMC article.
References
-
- Arias, A., E. Lázaro, C. Escarmís, and E. Domingo. 2001. Molecular intermediates of fitness gain of an RNA virus: characterization of a mutant spectrum by biological and molecular cloning. J. Gen. Virol. 82:1049-1060. - PubMed
-
- Ayllón, M. A., C. López, J. Navas-Castillo, S. M. Garnsey, J. Guerri, R. Flores, and P. Moreno. 2001. Polymorphism of the 5′ terminal region of Citrus tristeza virus (CTV) RNA: incidence of three sequence types in isolates of different origin and pathogenicity. Arch. Virol. 146:27-40. - PubMed
-
- Ayllón, M. A., C. López, J. Navas-Castillo, M. Mawassi, W. O. Dawson, J. Guerri, R. Flores, and P. Moreno. 1999. New defective RNAs from Citrus tristeza virus: evidence for a replicase driven template switching mechanism in their generation. J. Gen. Virol. 80:817-821. - PubMed
-
- Chao, L. 1988. Evolution of sex in RNA viruses. J. Theor. Biol. 133:99-112. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

