Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication
- PMID: 19793813
- PMCID: PMC2786852
- DOI: 10.1128/JVI.00942-09
Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication
Abstract
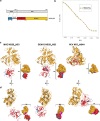
The flavivirus genome comprises a single strand of positive-sense RNA, which is translated into a polyprotein and cleaved by a combination of viral and host proteases to yield functional proteins. One of these, nonstructural protein 3 (NS3), is an enzyme with both serine protease and NTPase/helicase activities. NS3 plays a central role in the flavivirus life cycle: the NS3 N-terminal serine protease together with its essential cofactor NS2B is involved in the processing of the polyprotein, whereas the NS3 C-terminal NTPase/helicase is responsible for ATP-dependent RNA strand separation during replication. An unresolved question remains regarding why NS3 appears to encode two apparently disconnected functionalities within one protein. Here we report the 2.75-A-resolution crystal structure of full-length Murray Valley encephalitis virus NS3 fused with the protease activation peptide of NS2B. The biochemical characterization of this construct suggests that the protease has little influence on the helicase activity and vice versa. This finding is in agreement with the structural data, revealing a single protein with two essentially segregated globular domains. Comparison of the structure with that of dengue virus type 4 NS2B-NS3 reveals a relative orientation of the two domains that is radically different between the two structures. Our analysis suggests that the relative domain-domain orientation in NS3 is highly variable and dictated by a flexible interdomain linker. The possible implications of this conformational flexibility for the function of NS3 are discussed.
Figures




References
-
- Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948-1954. - PubMed
-
- Arias, C. F., F. Preugschat, and J. H. Strauss. 1993. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology 193:888-899. - PubMed
-
- Assenberg, R., J. Ren, A. Verma, T. S. Walter, D. Alderton, R. J. Hurrelbrink, S. D. Fuller, S. Bressanelli, R. J. Owens, D. I. Stuart, and J. M. Grimes. 2007. Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues. J. Gen. Virol. 88:2228-2236. - PubMed
-
- Benarroch, D., B. Selisko, G. A. Locatelli, G. Maga, J. L. Romette, and B. Canard. 2004. The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology 328:208-218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

