Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein
- PMID: 19793905
- PMCID: PMC2885062
- DOI: 10.1099/vir.0.015909-0
Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein
Abstract
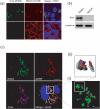
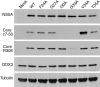
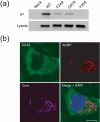
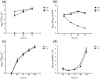
The cellular DEAD-box protein DDX3 was recently shown to be essential for hepatitis C virus (HCV) replication. Prior to that, we had reported that HCV core binds to DDX3 in yeast-two hybrid and transient transfection assays. Here, we confirm by co-immunoprecipitation that this interaction occurs in cells replicating the JFH1 virus. Consistent with this result, immunofluorescence staining of infected cells revealed a dramatic redistribution of cytoplasmic DDX3 by core protein to the virus assembly sites around lipid droplets. Given this close association of DDX3 with core and lipid droplets, and its involvement in virus replication, we investigated the importance of this host factor in the virus life cycle. Mutagenesis studies located a single amino acid in the N-terminal domain of JFH1 core that when changed to alanine significantly abrogated this interaction. Surprisingly, this mutation did not alter infectious virus production and RNA replication, indicating that the core-DDX3 interaction is dispensable in the HCV life cycle. Consistent with previous studies, siRNA-led knockdown of DDX3 lowered virus production and RNA replication levels of both WT JFH1 and the mutant virus unable to bind DDX3. Thus, our study shows for the first time that the requirement of DDX3 for HCV replication is unrelated to its interaction with the viral core protein.
Figures


Similar articles
-
DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication.J Virol. 2007 Dec;81(24):13922-6. doi: 10.1128/JVI.01517-07. Epub 2007 Sep 12. J Virol. 2007. PMID: 17855521 Free PMC article.
-
Hepatitis C virus core protein abrogates the DDX3 function that enhances IPS-1-mediated IFN-beta induction.PLoS One. 2010 Dec 8;5(12):e14258. doi: 10.1371/journal.pone.0014258. PLoS One. 2010. PMID: 21170385 Free PMC article.
-
Hepatitis C virus core protein interacts with a human DEAD box protein DDX3.Virology. 1999 May 10;257(2):330-40. doi: 10.1006/viro.1999.9659. Virology. 1999. PMID: 10329544
-
DEAD-box RNA Helicase DDX3: Functional Properties and Development of DDX3 Inhibitors as Antiviral and Anticancer Drugs.Molecules. 2020 Feb 24;25(4):1015. doi: 10.3390/molecules25041015. Molecules. 2020. PMID: 32102413 Free PMC article. Review.
-
Understanding the interaction of hepatitis C virus with host DEAD-box RNA helicases.World J Gastroenterol. 2014 Mar 21;20(11):2913-26. doi: 10.3748/wjg.v20.i11.2913. World J Gastroenterol. 2014. PMID: 24659882 Free PMC article. Review.
Cited by
-
Hepatitis C virus core-derived peptides inhibit genotype 1b viral genome replication via interaction with DDX3X.PLoS One. 2010 Sep 17;5(9):e12826. doi: 10.1371/journal.pone.0012826. PLoS One. 2010. PMID: 20862261 Free PMC article.
-
Global Effects of DDX3 Inhibition on Cell Cycle Regulation Identified by a Combined Phosphoproteomics and Single Cell Tracking Approach.Transl Oncol. 2018 Jun;11(3):755-763. doi: 10.1016/j.tranon.2018.04.001. Epub 2018 Apr 24. Transl Oncol. 2018. PMID: 29684792 Free PMC article.
-
Signals Involved in Regulation of Hepatitis C Virus RNA Genome Translation and Replication.Front Microbiol. 2018 Mar 12;9:395. doi: 10.3389/fmicb.2018.00395. eCollection 2018. Front Microbiol. 2018. PMID: 29593672 Free PMC article. Review.
-
The ATP-Dependent RNA Helicase DDX3X Modulates Herpes Simplex Virus 1 Gene Expression.J Virol. 2017 Mar 29;91(8):e02411-16. doi: 10.1128/JVI.02411-16. Print 2017 Apr 15. J Virol. 2017. PMID: 28148788 Free PMC article.
-
The DEAD-box helicase DDX3 supports the assembly of functional 80S ribosomes.Nucleic Acids Res. 2012 Jun;40(11):4998-5011. doi: 10.1093/nar/gks070. Epub 2012 Feb 9. Nucleic Acids Res. 2012. PMID: 22323517 Free PMC article.
References
-
- Barba, G., Harper, F., Harada, T., Kohara, M., Goulinet, S., Matsuura, Y., Eder, G., Schaff, Z., Chapman, M. J. & other authors (1997). Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci U S A 94, 1200–1205. - PMC - PubMed
-
- Blight, K. J., Kolykhalov, A. A. & Rice, C. M. (2000). Efficient initiation of HCV RNA replication in cell culture. Science 290, 1972–1974. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

