DNA microarray-based identification and typing of Actinobacillus pleuropneumoniae
- PMID: 19794891
- PMCID: PMC2705073
DNA microarray-based identification and typing of Actinobacillus pleuropneumoniae
Abstract
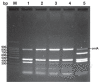
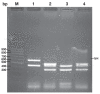
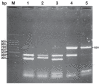
A DNA microarray system was prepared and shown to facilitate identification and typing of Actinobacillus pleuropneumoniae. The DNA microarray, composed of 18 DNA polymerase chain reaction (PCR) amplicons printed on glass slides and arranged in 3 subarrays, was developed. These target DNA included 1 or multiple fragments of the outer membrane lipoprotein, apx toxin, capsular polysaccharide, and disulfide bound formation protein E (dsbE)-like genes of A. pleuropneumoniae. These arrayed target DNA retained their expected hybridization properties. The hybridization signal intensities ranged from the least-intense to the most-intense, 4626 to 9789 arbitrary fluorescence units, respectively. Cy3-probes of A. pleuropneumoniae strains labeled with multiplex PCR were hybridized to the DNA microarray. A total of 51 different A. pleuropneumoniae strains representing serotype 1 to 12 reference strains and clinical isolates were detected and typed by the DNA microarray. Twelve reference serotypes produced 11 distinct target DNA hybridization patterns, and hybridization patterns of serotypes 1 (n = 7), 3 (n = 5), and 7 (n = 6) field isolates were identical to hybridization patterns of reference serotypes 1, 3, and 7, respectively. Non-serotyped isolates 4, 6, and 11 (out of 21) from diseased pigs had identical hybridization patterns to reference serotypes 3, 7, and 1, respectively. The results show that the DNA microarray system described in the present study is a valuable tool for identifying and typing reference strains and isolates of A. pleuropneumoniae, and enables relatively rapid identification of non-serotyped isolates.
Une puce à ADN a été préparée et a facilité l’identification et le typage d’Actinobacillus pleuropneumoniae. La puce à ADN a été développée et était composée de 18 amplicons d’ADN obtenus par réaction d’amplification en chaîne par la polymérase (PCR) imprimés sur des lames de verre et arrangés en 3 modules. Les ADN cibles incluaient des fragments uniques ou multiples des gènes de la lipoprotéine de membrane externe, la toxine apx, le polysaccharide capsulaire et une protéine apparentée à la protéine E pour la formation des ponts disulfures (dsbE) d’Actionbacillus pleuropneumoniae. Ces ADN cibles ont conservé leurs propriétés d’hybridation attendues. L’intensité des signaux d’hybridation variait du moins intense au plus intense, respectivement 4626 à 9789 unités fluorescentes arbitraires. Des sondes Cy3 de souches d’A. pleuropneumoniae marquées avec un PCR multiplex ont été hybridées à la puce ADN. Un total de 51 souches différentes d’A. pleuropneumoniae représentant les souches de références des sérotypes 1 à 12 et des isolats cliniques ont été détectées et typées par la puce à ADN. Les douze sérotypes de référence ont donné 11 patrons d’hybridation spécifiques des ADN cibles, et les patrons d’hybridation d’isolats cliniques des sérotypes 1 (n = 7), 3 (n = 5) et 7 (n = 6) étaient identiques aux patrons d’hybridation des souches de référence des sérotypes 1, 3 et 7, respectivement. Sur un total de 21 isolats non-sérotypés provenant de porcs malades, 4, 6 et 11 avaient des patrons d’hybridation identiques, respectivement, aux sérotypes 3, 7 et 1. Les résultats démontrent que le système de puce à ADN décrit dans la présente étude est un outil valide pour identifier et type des souches de référence et des isolats d’A. pleuropneumoniae, et permet une identification relativement rapide des isolats non-sérotypables.
(Traduit par Docteur Serge Messier)
Figures



Similar articles
-
An Actinobacillus pleuropneumoniae PCR typing system based on the apx and omlA genes--evaluation of isolates from lungs and tonsils of pigs.Vet Microbiol. 2000 Jul 3;75(1):43-57. doi: 10.1016/s0378-1135(00)00206-6. Vet Microbiol. 2000. PMID: 10865151
-
PCR-based identification of serotype 2 isolates of Actinobacillus pleuropneumoniae biovars I and II.Vet Microbiol. 2004 Apr 19;99(3-4):307-10. doi: 10.1016/j.vetmic.2003.12.007. Vet Microbiol. 2004. PMID: 15066734
-
Detection and identification of Actinobacillus pleuropneumoniae serotype 5 by multiplex PCR.J Clin Microbiol. 1998 Jun;36(6):1704-10. doi: 10.1128/JCM.36.6.1704-1710.1998. J Clin Microbiol. 1998. PMID: 9620404 Free PMC article.
-
Detection, identification, and subtyping of Actinobacillus pleuropneumoniae.Methods Mol Biol. 2003;216:87-95. doi: 10.1385/1-59259-344-5:87. Methods Mol Biol. 2003. PMID: 12512357 Review. No abstract available.
-
The challenge of detecting herds sub-clinically infected with Actinobacillus pleuropneumoniae.Vet J. 2015 Oct;206(1):30-8. doi: 10.1016/j.tvjl.2015.06.016. Epub 2015 Jul 2. Vet J. 2015. PMID: 26206322 Review.
Cited by
-
Explorative Field Study on the Use of Oral Fluids for the Surveillance of Actinobacillus pleuropneumoniae Infections in Fattening Farms by an Apx-Real-Time PCR.Vet Sci. 2022 Oct 8;9(10):552. doi: 10.3390/vetsci9100552. Vet Sci. 2022. PMID: 36288165 Free PMC article.
References
-
- Taylor DJ. Actinobacillus pleuropneumoniae. In: Leman AD, Straw BE, Mengeling WL, et al., editors. Diseases of Swine. 8th ed. Ames, Iowa: Iowa State University Pr; 1999. pp. 343–354.
-
- Blackall PJ, Klaasen HL, van den Bosch H, Kuhnert P, Frey J. Proposal of a new serovar of Actinobacillus pleuropneumoniae: serovar 15. Vet Microbiol. 2002;84:47–52. - PubMed
-
- Bosse JT, Janson H, Sheehan BJ, et al. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. 2002;4:225–235. - PubMed
-
- Haesebrouck F, Chiers K, Van Overbeke I, Ducatelle R. Actinobacillus pleuropneumoniae infections in pigs: The role of virulence factors in pathogenesis and protection. Vet Microbiol. 1997;58:239–249. - PubMed
-
- Jacobsen MJ, Nielsen JP, Nielsen R. Comparison of virulence of different Actinobacillus pleuropneumoniae serotypes and biotypes using an aerosol infection model. Vet Microbiol. 1996;49:159–168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
