Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF
- PMID: 19794910
- PMCID: PMC2748705
- DOI: 10.1371/journal.pone.0007243
Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF
Abstract
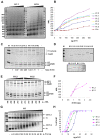
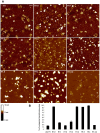
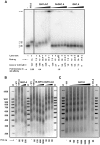
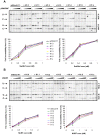
Although ubiquitously present in chromatin, the function of the linker histone subtypes is partly unknown and contradictory studies on their properties have been published. To explore whether the various H1 subtypes have a differential role in the organization and dynamics of chromatin we have incorporated all of the somatic human H1 subtypes into minichromosomes and compared their influence on nucleosome spacing, chromatin compaction and ATP-dependent remodeling. H1 subtypes exhibit different affinities for chromatin and different abilities to promote chromatin condensation, as studied with the Atomic Force Microscope. According to this criterion, H1 subtypes can be classified as weak condensers (H1.1 and H1.2), intermediate condensers (H1.3) and strong condensers (H1.0, H1.4, H1.5 and H1x). The variable C-terminal domain is required for nucleosome spacing by H1.4 and is likely responsible for the chromatin condensation properties of the various subtypes, as shown using chimeras between H1.4 and H1.2. In contrast to previous reports with isolated nucleosomes or linear nucleosomal arrays, linker histones at a ratio of one per nucleosome do not preclude remodeling of minichromosomes by yeast SWI/SNF or Drosophila NURF. We hypothesize that the linker histone subtypes are differential organizers of chromatin, rather than general repressors.
Conflict of interest statement
Figures

Similar articles
-
Solution AFM studies of human Swi-Snf and its interactions with MMTV DNA and chromatin.Biophys J. 2005 Nov;89(5):3386-98. doi: 10.1529/biophysj.105.065391. Epub 2005 Aug 12. Biophys J. 2005. PMID: 16100261 Free PMC article.
-
The core histone N-terminal domains are required for multiple rounds of catalytic chromatin remodeling by the SWI/SNF and RSC complexes.Biochemistry. 1999 Feb 23;38(8):2514-22. doi: 10.1021/bi982109d. Biochemistry. 1999. PMID: 10029546
-
Distortion of histone octamer core promotes nucleosome mobilization by a chromatin remodeler.Science. 2017 Jan 20;355(6322):eaaa3761. doi: 10.1126/science.aaa3761. Science. 2017. PMID: 28104838 Free PMC article.
-
The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle.EMBO Rep. 2015 Nov;16(11):1439-53. doi: 10.15252/embr.201540749. Epub 2015 Oct 15. EMBO Rep. 2015. PMID: 26474902 Free PMC article. Review.
-
Linker histones in hormonal gene regulation.Biochim Biophys Acta. 2016 Mar;1859(3):520-5. doi: 10.1016/j.bbagrm.2015.10.016. Epub 2015 Oct 27. Biochim Biophys Acta. 2016. PMID: 26518266 Review.
Cited by
-
Regulation of Cellular Dynamics and Chromosomal Binding Site Preference of Linker Histones H1.0 and H1.X.Mol Cell Biol. 2016 Oct 13;36(21):2681-2696. doi: 10.1128/MCB.00200-16. Print 2016 Nov 1. Mol Cell Biol. 2016. PMID: 27528617 Free PMC article.
-
Developmentally regulated linker histone H1c promotes heterochromatin condensation and mediates structural integrity of rod photoreceptors in mouse retina.J Biol Chem. 2013 Jun 14;288(24):17895-907. doi: 10.1074/jbc.M113.452144. Epub 2013 May 3. J Biol Chem. 2013. PMID: 23645681 Free PMC article.
-
Use of biotinylated plasmid DNA as a surrogate for HSV DNA to identify proteins that repress or activate viral gene expression.Proc Natl Acad Sci U S A. 2012 Dec 18;109(51):E3549-57. doi: 10.1073/pnas.1218783109. Epub 2012 Dec 5. Proc Natl Acad Sci U S A. 2012. PMID: 23223531 Free PMC article.
-
Heterologous expression of bovine histone H1foo into porcine fibroblasts alters the transcriptome profile but not embryo development following nuclear transfer.J Assist Reprod Genet. 2025 Apr;42(4):1109-1120. doi: 10.1007/s10815-025-03437-1. Epub 2025 Mar 3. J Assist Reprod Genet. 2025. PMID: 40025368
-
High-resolution mapping of h1 linker histone variants in embryonic stem cells.PLoS Genet. 2013;9(4):e1003417. doi: 10.1371/journal.pgen.1003417. Epub 2013 Apr 25. PLoS Genet. 2013. PMID: 23633960 Free PMC article.
References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
-
- Ramakrishnan V. Histone structure and the organization of the nucleosome. Annu Rev Biophys Biomol Struct. 1997;26:83–112. - PubMed
-
- Huynh VA, Robinson PJ, Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol. 2005;345:957–968. - PubMed
-
- Butler PJ, Thomas JO. Changes in chromatin folding in solution. J Mol Biol. 1980;140:505–529. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

