Inferring the transcriptional landscape of bovine skeletal muscle by integrating co-expression networks
- PMID: 19794913
- PMCID: PMC2749936
- DOI: 10.1371/journal.pone.0007249
Inferring the transcriptional landscape of bovine skeletal muscle by integrating co-expression networks
Abstract
Background: Despite modern technologies and novel computational approaches, decoding causal transcriptional regulation remains challenging. This is particularly true for less well studied organisms and when only gene expression data is available. In muscle a small number of well characterised transcription factors are proposed to regulate development. Therefore, muscle appears to be a tractable system for proposing new computational approaches.
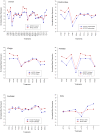
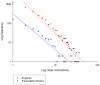
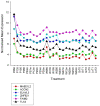
Methodology/principal findings: Here we report a simple algorithm that asks "which transcriptional regulator has the highest average absolute co-expression correlation to the genes in a co-expression module?" It correctly infers a number of known causal regulators of fundamental biological processes, including cell cycle activity (E2F1), glycolysis (HLF), mitochondrial transcription (TFB2M), adipogenesis (PIAS1), neuronal development (TLX3), immune function (IRF1) and vasculogenesis (SOX17), within a skeletal muscle context. However, none of the canonical pro-myogenic transcription factors (MYOD1, MYOG, MYF5, MYF6 and MEF2C) were linked to muscle structural gene expression modules. Co-expression values were computed using developing bovine muscle from 60 days post conception (early foetal) to 30 months post natal (adulthood) for two breeds of cattle, in addition to a nutritional comparison with a third breed. A number of transcriptional landscapes were constructed and integrated into an always correlated landscape. One notable feature was a 'metabolic axis' formed from glycolysis genes at one end, nuclear-encoded mitochondrial protein genes at the other, and centrally tethered by mitochondrially-encoded mitochondrial protein genes.
Conclusions/significance: The new module-to-regulator algorithm complements our recently described Regulatory Impact Factor analysis. Together with a simple examination of a co-expression module's contents, these three gene expression approaches are starting to illuminate the in vivo transcriptional regulation of skeletal muscle development.
Conflict of interest statement
Figures




References
-
- Takebe A, Era T, Okada M, Martin Jakt L, Kuroda Y, et al. Microarray analysis of PDGFR alpha+ populations in ES cell differentiation culture identifies genes involved in differentiation of mesoderm and mesenchyme including ARID3b that is essential for development of embryonic mesenchymal cells. Dev Biol. 2006;293:25–37. - PubMed
-
- Tuch BB, Li H, Johnson AD. Evolution of eukaryotic transcription circuits. Science. 2008;319:1797–1799. - PubMed
-
- Blais A, Dynlacht BD. Constructing transcriptional regulatory networks. Genes Dev. 2005;19:1499–1511. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

