KA-SB: from data integration to large scale reasoning
- PMID: 19796402
- PMCID: PMC2755826
- DOI: 10.1186/1471-2105-10-S10-S5
KA-SB: from data integration to large scale reasoning
Abstract
Background: The analysis of information in the biological domain is usually focused on the analysis of data from single on-line data sources. Unfortunately, studying a biological process requires having access to disperse, heterogeneous, autonomous data sources. In this context, an analysis of the information is not possible without the integration of such data.
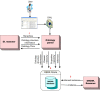
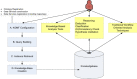
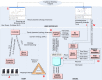
Methods: KA-SB is a querying and analysis system for final users based on combining a data integration solution with a reasoner. Thus, the tool has been created with a process divided into two steps: 1) KOMF, the Khaos Ontology-based Mediator Framework, is used to retrieve information from heterogeneous and distributed databases; 2) the integrated information is crystallized in a (persistent and high performance) reasoner (DBOWL). This information could be further analyzed later (by means of querying and reasoning).
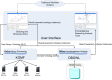
Results: In this paper we present a novel system that combines the use of a mediation system with the reasoning capabilities of a large scale reasoner to provide a way of finding new knowledge and of analyzing the integrated information from different databases, which is retrieved as a set of ontology instances. This tool uses a graphical query interface to build user queries easily, which shows a graphical representation of the ontology and allows users o build queries by clicking on the ontology concepts.
Conclusion: These kinds of systems (based on KOMF) will provide users with very large amounts of information (interpreted as ontology instances once retrieved), which cannot be managed using traditional main memory-based reasoners. We propose a process for creating persistent and scalable knowledgebases from sets of OWL instances obtained by integrating heterogeneous data sources with KOMF. This process has been applied to develop a demo tool http://khaos.uma.es/KA-SB, which uses the BioPax Level 3 ontology as the integration schema, and integrates UNIPROT, KEGG, CHEBI, BRENDA and SABIORK databases.
Figures




References
-
- Risch T, Josifovski V. Distributed data integration by object-oriented mediator servers. Concurrency and Computation: Practice and Experience. 2001;14:1–21.
-
- Tomasic A, Amouroux R, Bonnet P, Kapitskaia O, Naacke H, Raschid L. Proceedings of the 1997ACM SIGMOD International Conference on Management of Data: 11–15 May 1997; New York. 1997. The distributed information search component (disco) and the world wide web; pp. 543–545.
-
- Garcia-Molina H, Papakonstantinou Y, Quass D, Rajaraman A, Sagiv Y, Ullman J, Vassalos V, Widom J. The tsimmis aproach to mediation: Data models and languages. Journal of Intelligent Information Systems. 1997;8(2):117–132. doi: 10.1023/A:1008683107812. - DOI
-
- Haas L, Kossmann D, Wimmers E, Yang J. An optimizer for heterogeneous systems with nonstandard data and search capabilities. Data Engineering Bulletin. 1996;19:37–44.
-
- Ksiezyk T, Martin G, Jia Q. Proceedings of the 25th International Computer Software and Applications Conference on Invigorating Software Development; 8–12 October 2001; Chicago. 2001. Infosleuth: Agent-based system for data integration and analysis; p. 474.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

