SynCAM1 recruits NMDA receptors via protein 4.1B
- PMID: 19796685
- PMCID: PMC2784006
- DOI: 10.1016/j.mcn.2009.09.010
SynCAM1 recruits NMDA receptors via protein 4.1B
Abstract
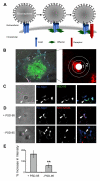
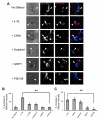
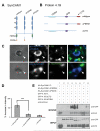
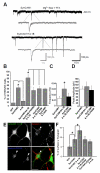
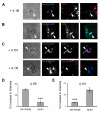
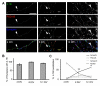
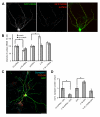
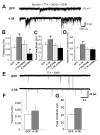
Cell adhesion molecules have been implicated as key organizers of synaptic structures, but there is still a need to determine how these molecules facilitate neurotransmitter receptor recruitment to developing synapses. Here, we identify erythrocyte protein band 4.1-like 3 (protein 4.1B) as an intracellular effector molecule of Synaptic Cell Adhesion Molecule 1 (SynCAM1) that is sufficient to recruit NMDA-type receptors (NMDARs) to SynCAM1 adhesion sites in COS7 cells. Protein 4.1B in conjunction with SynCAM1 also increased the frequency of NMDAR-mediated mEPSCs and area of presynaptic contact in an HEK293 cell/ neuron co-culture assay. Studies in cultured hippocampal neurons reveal that manipulation of protein 4.1B expression levels specifically affects NMDAR-mediated activity and localization. Finally, further experimentation in COS7 cells show that SynCAM1 may also interact with protein 4.1N to specifically effect AMPA type receptor (AMPAR) recruitment. Thus, SynCAM1 may recruit both AMPARs and NMDARs by independent mechanisms during synapse formation.
Figures
Similar articles
-
SynCAM1 expression correlates with restoration of central synapses on spinal motoneurons after two different models of peripheral nerve injury.J Comp Neurol. 2009 Dec 10;517(5):670-82. doi: 10.1002/cne.22186. J Comp Neurol. 2009. PMID: 19827159
-
Neuroligin1: a cell adhesion molecule that recruits PSD-95 and NMDA receptors by distinct mechanisms during synaptogenesis.Neural Dev. 2009 May 18;4:17. doi: 10.1186/1749-8104-4-17. Neural Dev. 2009. PMID: 19450252 Free PMC article.
-
Neuronal RA175/SynCAM1 isoforms are processed by tumor necrosis factor-alpha-converting enzyme (TACE)/ADAM17-like proteases.Neurosci Lett. 2008 Oct 17;444(1):16-21. doi: 10.1016/j.neulet.2008.08.023. Epub 2008 Aug 13. Neurosci Lett. 2008. PMID: 18718504
-
Diminished neuronal activity increases neuron-neuron connectivity underlying silent synapse formation and the rapid conversion of silent to functional synapses.J Neurosci. 2005 Apr 20;25(16):4040-51. doi: 10.1523/JNEUROSCI.4115-04.2005. J Neurosci. 2005. PMID: 15843606 Free PMC article.
-
Regulation of neuronal PKA signaling through AKAP targeting dynamics.Eur J Cell Biol. 2006 Jul;85(7):627-33. doi: 10.1016/j.ejcb.2006.01.010. Epub 2006 Feb 28. Eur J Cell Biol. 2006. PMID: 16504338 Review.
Cited by
-
SynCAMs in Normal Vertebrate Neural Development and Neuropsychiatric Disorders: from the Perspective of the OCAs.Mol Neurobiol. 2024 Jan;61(1):358-371. doi: 10.1007/s12035-023-03579-2. Epub 2023 Aug 22. Mol Neurobiol. 2024. PMID: 37607992 Review.
-
CADM1 controls actin cytoskeleton assembly and regulates extracellular matrix adhesion in human mast cells.PLoS One. 2014 Jan 22;9(1):e85980. doi: 10.1371/journal.pone.0085980. eCollection 2014. PLoS One. 2014. PMID: 24465823 Free PMC article.
-
Changes in nucleus accumbens core translatome accompanying incubation of cocaine craving.Neuropsychopharmacology. 2025 Jul;50(8):1305-1316. doi: 10.1038/s41386-025-02112-4. Epub 2025 Apr 29. Neuropsychopharmacology. 2025. PMID: 40301580
-
The 4.1B cytoskeletal protein regulates the domain organization and sheath thickness of myelinated axons.Glia. 2013 Feb;61(2):240-53. doi: 10.1002/glia.22430. Epub 2012 Oct 26. Glia. 2013. PMID: 23109359 Free PMC article.
-
Immunoglobulin-Like Receptors and Their Impact on Wiring of Brain Synapses.Annu Rev Genet. 2018 Nov 23;52:567-590. doi: 10.1146/annurev-genet-120417-031513. Epub 2018 Sep 13. Annu Rev Genet. 2018. PMID: 30212237 Free PMC article. Review.
References
-
- An XL, Takakuwa Y, Nunomura W, Manno S, Mohandas N. Modulation of band 3-ankyrin interaction by protein 4.1. Functional implications in regulation of erythrocyte membrane mechanical properties. J Biol Chem. 1996;271:33187–33191. - PubMed
-
- Biederer T. Bioinformatic characterization of the SynCAM family of immunoglobulin-like domain-containing adhesion molecules. Genomics. 2005a - PubMed
-
- Biederer T. Progress from the postsynaptic side: signaling in synaptic differentiation. Sci STKE. 2005b;2005:pe9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

