LGI1-associated epilepsy through altered ADAM23-dependent neuronal morphology
- PMID: 19796686
- PMCID: PMC2783222
- DOI: 10.1016/j.mcn.2009.09.008
LGI1-associated epilepsy through altered ADAM23-dependent neuronal morphology
Abstract
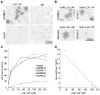
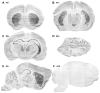
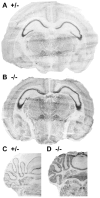
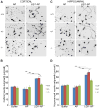
Most epilepsy genes encode ion channels, but the LGI1 gene responsible for autosomal dominant partial epilepsy with auditory features produces a secreted protein. LGI1 is suggested to regulate PSD-95 via ADAM22. However, no unbiased screen of LGI1 action has been conducted. Here, we searched for brain genes supporting high affinity LGI-1 binding. ADAM23 was the only LGI1 interactor identified. The related proteins, ADAM22 and ADAM11, but not ADAM12, bind LGI1. Neither ADAM23 nor ADAM11, nor some forms of ADAM22, contain PDZ-interacting sequences, suggesting PSD-95-independent mechanisms in ADPEAF. Because ADAMs modulate integrins, we examined LGI1 effect on neurite outgrowth. LGI1 increases outgrowth from wild-type but not ADAM23-/- neurons. Furthermore, CA1 pyramidal neurons of ADAM23-/- hippocampi have reduced dendritic arborization. ADAM23-/- mice exhibit spontaneous seizures, while ADAM23+/- mice have decreased seizure thresholds. Thus, LGI1 binding to ADAM23 is necessary to correctly pattern neuronal morphology and altered anatomical patterning contributes to ADPEAF.
Figures



Similar articles
-
Secretion-Positive LGI1 Mutations Linked to Lateral Temporal Epilepsy Impair Binding to ADAM22 and ADAM23 Receptors.PLoS Genet. 2016 Oct 19;12(10):e1006376. doi: 10.1371/journal.pgen.1006376. eCollection 2016 Oct. PLoS Genet. 2016. PMID: 27760137 Free PMC article.
-
LGI1 and LGI4 bind to ADAM22, ADAM23 and ADAM11.Int J Biol Sci. 2008;4(6):387-96. doi: 10.7150/ijbs.4.387. Epub 2008 Oct 21. Int J Biol Sci. 2008. PMID: 18974846 Free PMC article.
-
Autoantibodies to epilepsy-related LGI1 in limbic encephalitis neutralize LGI1-ADAM22 interaction and reduce synaptic AMPA receptors.J Neurosci. 2013 Nov 13;33(46):18161-74. doi: 10.1523/JNEUROSCI.3506-13.2013. J Neurosci. 2013. PMID: 24227725 Free PMC article.
-
Insights into the mechanisms of epilepsy from structural biology of LGI1-ADAM22.Cell Mol Life Sci. 2020 Jan;77(2):267-274. doi: 10.1007/s00018-019-03269-0. Epub 2019 Aug 20. Cell Mol Life Sci. 2020. PMID: 31432233 Free PMC article. Review.
-
LGI1: from zebrafish to human epilepsy.Prog Brain Res. 2014;213:159-79. doi: 10.1016/B978-0-444-63326-2.00009-0. Prog Brain Res. 2014. PMID: 25194489 Review.
Cited by
-
The LGI1-ADAM22 protein complex directs synapse maturation through regulation of PSD-95 function.Proc Natl Acad Sci U S A. 2015 Jul 28;112(30):E4129-37. doi: 10.1073/pnas.1511910112. Epub 2015 Jul 15. Proc Natl Acad Sci U S A. 2015. PMID: 26178195 Free PMC article.
-
Electroclinical characterization of epileptic seizures in leucine-rich, glioma-inactivated 1-deficient mice.Brain. 2010 Sep;133(9):2749-62. doi: 10.1093/brain/awq171. Epub 2010 Jul 21. Brain. 2010. PMID: 20659958 Free PMC article.
-
Pathophysiology of Trans-Synaptic Adhesion Molecules: Implications for Epilepsy.Front Cell Dev Biol. 2018 Sep 21;6:119. doi: 10.3389/fcell.2018.00119. eCollection 2018. Front Cell Dev Biol. 2018. PMID: 30298130 Free PMC article. Review.
-
LGI2 truncation causes a remitting focal epilepsy in dogs.PLoS Genet. 2011 Jul;7(7):e1002194. doi: 10.1371/journal.pgen.1002194. Epub 2011 Jul 28. PLoS Genet. 2011. PMID: 21829378 Free PMC article.
-
Chemical corrector treatment ameliorates increased seizure susceptibility in a mouse model of familial epilepsy.Nat Med. 2015 Jan;21(1):19-26. doi: 10.1038/nm.3759. Epub 2014 Dec 8. Nat Med. 2015. PMID: 25485908
References
-
- Bisulli F, Tinuper P, Avoni P, Striano P, Striano S, d'Orsi G, Vignatelli L, Bagattin A, Scudellaro E, Florindo I, Nobile C, Tassinari CA, Baruzzi A, Michelucci R. Idiopathic partial epilepsy with auditory features (IPEAF): a clinical and genetic study of 53 sporadic cases. Brain. 2004;127:1343–1352. - PubMed
-
- Chabrol E, Gourfinkel-An I, Scheffer IE, Picard F, Couarch P, Berkovic SF, McMahon JM, Bajaj N, Mota-Vieira L, Mota R, Trouillard O, Depienne C, Baulac M, LeGuern E, Baulac S. Absence of mutations in the LGI1 receptor ADAM22 gene in autosomal dominant lateral temporal epilepsy. Epilepsy Res. 2007;76:41–48. - PubMed
-
- Diani E, et al. Autosomal dominant lateral temporal epilepsy: absence of mutations in ADAM22 and Kv1 channel genes encoding LGI1-associated proteins. Epilepsy Res. 2008;80:1–8. - PubMed
-
- Fertig E, Lincoln A, Martinuzzi A, Mattson RH, Hisama FM. Novel LGI1 mutation in a family with autosomal dominant partial epilepsy with auditory features. Neurology. 2003;60:1687–1690. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

