Energetics, epigenetics, mitochondrial genetics
- PMID: 19796712
- PMCID: PMC3245717
- DOI: 10.1016/j.mito.2009.09.006
Energetics, epigenetics, mitochondrial genetics
Abstract
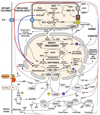
The epigenome has been hypothesized to provide the interface between the environment and the nuclear DNA (nDNA) genes. Key factors in the environment are the availability of calories and demands on the organism's energetic capacity. Energy is funneled through glycolysis and mitochondrial oxidative phosphorylation (OXPHOS), the cellular bioenergetic systems. Since there are thousands of bioenergetic genes dispersed across the chromosomes and mitochondrial DNA (mtDNA), both cis and trans regulation of the nDNA genes is required. The bioenergetic systems convert environmental calories into ATP, acetyl-Coenzyme A (acetyl-CoA), s-adenosyl-methionine (SAM), and reduced NAD(+). When calories are abundant, ATP and acetyl-CoA phosphorylate and acetylate chromatin, opening the nDNA for transcription and replication. When calories are limiting, chromatin phosphorylation and acetylation are lost and gene expression is suppressed. DNA methylation via SAM can also be modulated by mitochondrial function. Phosphorylation and acetylation are also pivotal to regulating cellular signal transduction pathways. Therefore, bioenergetics provides the interface between the environment and the epigenome. Consistent with this conclusion, the clinical phenotypes of bioenergetic diseases are strikingly similar to those observed in epigenetic diseases (Angelman, Rett, Fragile X Syndromes, the laminopathies, cancer, etc.), and an increasing number of epigenetic diseases are being associated with mitochondrial dysfunction. This bioenergetic-epigenomic hypothesis has broad implications for the etiology, pathophysiology, and treatment of a wide range of common diseases.
Figures
References
-
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. - PubMed
-
- Ahmed S, Passos JF, Birket MJ, Beckmann T, Brings S, Peters H, Birch-Machin MA, von Zglinicki T, Saretzki G. Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. Journal of Cell Science. 2008;121:1046–1053. - PubMed
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of The Cell. New York and London: Garland Science; 2002.
-
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genetics. 1999;23:185–188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

