Phylodynamics of infectious disease epidemics
- PMID: 19797047
- PMCID: PMC2787429
- DOI: 10.1534/genetics.109.106021
Phylodynamics of infectious disease epidemics
Abstract
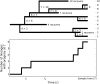
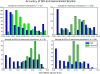
We present a formalism for unifying the inference of population size from genetic sequences and mathematical models of infectious disease in populations. Virus phylogenies have been used in many recent studies to infer properties of epidemics. These approaches rely on coalescent models that may not be appropriate for infectious diseases. We account for phylogenetic patterns of viruses in susceptible-infected (SI), susceptible-infected-susceptible (SIS), and susceptible-infected-recovered (SIR) models of infectious disease, and our approach may be a viable alternative to demographic models used to reconstruct epidemic dynamics. The method allows epidemiological parameters, such as the reproductive number, to be estimated directly from viral sequence data. We also describe patterns of phylogenetic clustering that are often construed as arising from a short chain of transmissions. Our model reproduces the moments of the distribution of phylogenetic cluster sizes and may therefore serve as a null hypothesis for cluster sizes under simple epidemiological models. We examine a small cross-sectional sample of human immunodeficiency (HIV)-1 sequences collected in the United States and compare our results to standard estimates of effective population size. Estimated prevalence is consistent with estimates of effective population size and the known history of the HIV epidemic. While our model accurately estimates prevalence during exponential growth, we find that periods of decline are harder to identify.
Figures




References
-
- Anderson, R. M., and R. M. May, 1991. Infectious Diseases of Humans: Dynamics and Control. Oxford University Press, London/New York/Oxford.
-
- Athreya, K. B., and P. E. Ney, 2004. Branching Processes. Dover, New York.
-
- Bailey, N. T. J., 1975. The Mathematical Theory of Infectious Diseases and Its Applications. Hafner Press, New York.
-
- Brenner, B. G., M. Roger, J. Routy, D. Moisi, M. Ntemgwa et al., 2007. High rates of forward transmission events after acute/early HIV-1 infection. J. Infect. Dis. 195 951. - PubMed
-
- Brown, A. J., H. F. Günthard, J. K. Wong, R. T. D'Aquila, V. A. Johnson et al., 1999. Sequence clusters in human immunodeficiency virus type 1 reverse transcriptase are associated with subsequent virological response to antiretroviral therapy. J. Infect. Dis. 180 1043–1049. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

