Total expression and dual gene-regulatory mechanisms maintained in deletions and duplications of the Pcdha cluster
- PMID: 19797050
- PMCID: PMC2797272
- DOI: 10.1074/jbc.M109.046938
Total expression and dual gene-regulatory mechanisms maintained in deletions and duplications of the Pcdha cluster
Abstract
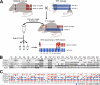
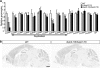
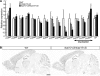
The clustered protocadherin-alpha (Pcdha) genes, which are expressed in the vertebrate brain, encode diverse membrane proteins whose functions are involved in axonal projection and in learning and memory. The Pcdha cluster consists of 14 tandemly arranged genes (Pcdha1-Pcdha12, Pcdhac1, and Pcdhac2, from 5' to 3'). Each first exon (the variable exons) is transcribed from its own promoter, and spliced to the constant exons, which are common to all the Pcdha genes. Cerebellar Purkinje cells show dual expression patterns for Pcdha. In individual Purkinje cells, different sets of the 5' genes in the cluster, Pcdha1-12, are randomly expressed, whereas both 3' genes, Pcdhac1 and Pcdhac2, are expressed constitutively. To elucidate the relationship between the genomic structure of the Pcdha cluster and their expression in Purkinje cells, we deleted or duplicated multiple variable exons and analyzed the expression of Pcdha genes in the mouse brain. In all mutant mice, transcript levels of the constant exons and the dual expression patterns were maintained. In the deletion mutants, the missing genes were flexibly compensated by the remaining variable exons. On the other hand, in duplication mutants, the levels of the duplicated genes were trimmed. These results indicate that the Pcdha genes are comprehensively regulated as a cluster unit, and that the regulators that randomly and constitutively drive Pcdha gene expression are intact in the deleted or duplicated mutant alleles. These dual regulatory mechanisms may play important roles in the diversity and fundamental functions of neurons.
Figures








References
-
- Imai T., Sakano H. (2008) Curr. Opin. Neurobiol. 18, 251–260 - PubMed
-
- Lomvardas S., Barnea G., Pisapia D. J., Mendelsohn M., Kirkland J., Axel R. (2006) Cell 126, 403–413 - PubMed
-
- Serafini T. (1999) Cell 98, 133–136 - PubMed
-
- Kohmura N., Senzaki K., Hamada S., Kai N., Yasuda R., Watanabe M., Ishii H., Yasuda M., Mishina M., Yagi T. (1998) Neuron 20, 1137–1151 - PubMed
-
- Esumi S., Kakazu N., Taguchi Y., Hirayama T., Sasaki A., Hirabayashi T., Koide T., Kitsukawa T., Hamada S., Yagi T. (2005) Nat. Genet. 37, 171–176 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

