BRCA1 localization to the telomere and its loss from the telomere in response to DNA damage
- PMID: 19797051
- PMCID: PMC2794724
- DOI: 10.1074/jbc.M109.025825
BRCA1 localization to the telomere and its loss from the telomere in response to DNA damage
Abstract
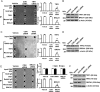
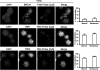
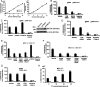
BRCA1, a tumor suppressor, participates in DNA damage signaling and repair. Previously, we showed that BRCA1 overexpression caused inhibition of telomerase activity and telomere shortening in breast and prostate cancer cells. We now report that BRCA1 knockdown causes increased telomerase reverse transcriptase expression, telomerase activity, and telomere length; but studies utilizing a combination of BRCA1 and telomerase reverse transcriptase small interfering RNAs suggest that BRCA1 also regulates telomere length independently of telomerase. Using telomeric chromatin immunoprecipitation assays, we detected BRCA1 at the telomere and demonstrated time-dependent loss of BRCA1 from the telomere following DNA damage. Further studies suggest that BRCA1 interacts with TRF1 and TRF2 in a DNA-dependent manner and that some of the nuclear BRCA1 colocalizes with TRF1/2. Our findings further suggest that Rad50 is required to localize BRCA1 at the telomere and that the association of BRCA1 with Rad50 does not require DNA. Finally, we found that BRCA1 regulates the length of the 3' G-rich overhang in a manner that is dependent upon Rad50. Our findings suggest that BRCA1 is recruited to the telomere in a Rad50-dependent manner and that BRCA1 may regulate telomere length and stability, in part through its presence at the telomere.
Figures
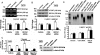






Similar articles
-
Expression of TRF1, TRF2, TIN2, TERT, KU70, and BRCA1 proteins is associated with telomere shortening and may contribute to multistage carcinogenesis of gastric cancer.J Cancer Res Clin Oncol. 2010 Sep;136(9):1407-14. doi: 10.1007/s00432-010-0795-x. Epub 2010 Feb 2. J Cancer Res Clin Oncol. 2010. PMID: 20127252 Free PMC article.
-
Expression of MRE11 complex (MRE11, RAD50, NBS1) and hRap1 and its relation with telomere regulation, telomerase activity in human gastric carcinomas.Pathobiology. 2001;69(4):219-24. doi: 10.1159/000055946. Pathobiology. 2001. PMID: 12007281
-
Coordinate regulation between expression levels of telomere-binding proteins and telomere length in breast carcinomas.Cancer Med. 2012 Oct;1(2):165-75. doi: 10.1002/cam4.14. Epub 2012 Jul 24. Cancer Med. 2012. PMID: 23342266 Free PMC article.
-
The Connection Between Cell Fate and Telomere.Adv Exp Med Biol. 2021;1275:71-100. doi: 10.1007/978-3-030-49844-3_3. Adv Exp Med Biol. 2021. PMID: 33539012 Review.
-
Regulation of telomerase by telomeric proteins.Annu Rev Biochem. 2004;73:177-208. doi: 10.1146/annurev.biochem.73.071403.160049. Annu Rev Biochem. 2004. PMID: 15189140 Review.
Cited by
-
BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage.Nat Commun. 2021 Jun 10;12(1):3542. doi: 10.1038/s41467-021-23716-6. Nat Commun. 2021. PMID: 34112789 Free PMC article.
-
Navigating the body of literature assessing BRCA1/2 mutations and markers of ovarian function: a systematic review and meta-analysis.J Assist Reprod Genet. 2020 May;37(5):1037-1055. doi: 10.1007/s10815-020-01745-2. Epub 2020 Mar 24. J Assist Reprod Genet. 2020. PMID: 32212026 Free PMC article.
-
Genetic Variations in Telomere Maintenance, with Implications on Tissue Renewal Capacity and Chronic Disease Pathologies.Curr Pharmacogenomics Person Med. 2010 Mar 1;8(1):7-24. doi: 10.2174/1875692111008010007. Curr Pharmacogenomics Person Med. 2010. PMID: 21258621 Free PMC article.
-
Antagonizing functions of BARD1 and its alternatively spliced variant BARD1δ in telomere stability.Oncotarget. 2017 Feb 7;8(6):9339-9353. doi: 10.18632/oncotarget.14068. Oncotarget. 2017. PMID: 28030839 Free PMC article.
-
Genetic susceptibility loci for subtypes of breast cancer in an African American population.Cancer Epidemiol Biomarkers Prev. 2013 Jan;22(1):127-34. doi: 10.1158/1055-9965.EPI-12-0769. Epub 2012 Nov 7. Cancer Epidemiol Biomarkers Prev. 2013. PMID: 23136140 Free PMC article.
References
-
- Morin G. B. (1989) Cell 59, 521–529 - PubMed
-
- Blackburn E. H. (1997) Biochemistry 62, 1196–1201 - PubMed
-
- McEachern M. J., Krauskopf A., Blackburn E. H. (2000) Annu. Rev. Genet. 34, 331–358 - PubMed
-
- Bodnar A. G., Ouellette M., Frolkis M., Holt S. E., Chiu C. P., Morin G. B., Harley C. B., Shay J. W., Lichtsteiner S., Wright W. E. (1998) Science 279, 349–352 - PubMed
-
- Greider C. W. (1999) Cell 97, 419–422 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

