Rhodococcus equi virulence-associated protein A is required for diversion of phagosome biogenesis but not for cytotoxicity
- PMID: 19797071
- PMCID: PMC2786453
- DOI: 10.1128/IAI.00856-09
Rhodococcus equi virulence-associated protein A is required for diversion of phagosome biogenesis but not for cytotoxicity
Abstract
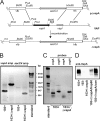
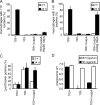
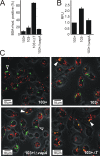
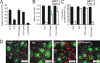
Rhodococcus equi is a gram-positive facultative intracellular pathogen that can cause severe bronchopneumonia in foals and AIDS patients. Virulence is plasmid regulated and is accompanied by phagosome maturation arrest and host cell necrosis. A replacement mutant in the gene for VapA (virulence-associated protein A), a major virulence factor of R. equi, was tested for its activities during macrophage infection. Early in infection, phagosomes containing the vapA mutant did not fuse with lysosomes and did not stain with the acidotropic fluor LysoTracker similar to those containing virulent wild-type R. equi. However, vapA mutant phagosomes had a lower average pH. Late in infection, phagosomes containing the vapA mutant were as frequently positive for LysoTracker as phagosomes containing plasmid-cured, avirulent bacteria, whereas those with virulent wild-type R. equi were still negative for the fluor. Macrophage necrosis after prolonged infection with virulent bacteria was accompanied by a loss of organelle staining with LysoTracker, suggesting that lysosome proton gradients had collapsed. The vapA mutant still killed the macrophages and yet did not affect the pH of host cell lysosomes. Hence, VapA is not required for host cell necrosis but is required for neutralization of phagosomes and lysosomes or their disruption. This is the first report of an R. equi mutant with altered phagosome biogenesis.
Figures
References
-
- Anes, E., M. P. Kuhnel, E. Bos, J. Moniz-Pereira, A. Habermann, and G. Griffiths. 2003. Selected lipids activate phagosome actin assembly and maturation resulting in killing of pathogenic mycobacteria. Nat. Cell Biol. 5:793-802. - PubMed
-
- Donisi, A., M. G. Suardi, S. Casari, M. Longo, G. P. Cadeo, and G. Carosi. 1996. Rhodococcus equi infection in HIV-infected patients. Aids 10:359-362. - PubMed
-
- Fernandez-Mora, E., M. Polidori, A. Luhrmann, U. E. Schaible, and A. Haas. 2005. Maturation of Rhodococcus equi-containing vacuoles is arrested after completion of the early endosome stage. Traffic 6:635-653. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

