Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0
- PMID: 19797078
- PMCID: PMC2753163
- DOI: 10.1083/jcb.200904110
Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0
Abstract
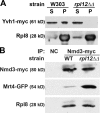
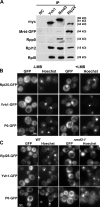
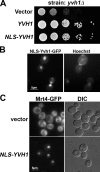
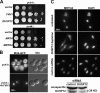
The ribosome stalk is essential for recruitment of translation factors. In yeast, P0 and Rpl12 correspond to bacterial L10 and L11 and form the stalk base of mature ribosomes, whereas Mrt4 is a nuclear paralogue of P0. In this study, we show that the dual-specificity phosphatase Yvh1 is required for the release of Mrt4 from the pre-60S subunits. Deletion of YVH1 leads to the persistence of Mrt4 on pre-60S subunits in the cytoplasm. A mutation in Mrt4 at the protein-RNA interface bypasses the requirement for Yvh1. Pre-60S subunits associated with Yvh1 contain Rpl12 but lack both Mrt4 and P0. These results suggest a linear series of events in which Yvh1 binds to the pre-60S subunit to displace Mrt4. Subsequently, P0 loads onto the subunit to assemble the mature stalk, and Yvh1 is released. The initial assembly of the ribosome with Mrt4 may provide functional compartmentalization of ribosome assembly in addition to the spatial separation afforded by the nuclear envelope.
Figures

Similar articles
-
Yvh1 is required for a late maturation step in the 60S biogenesis pathway.J Cell Biol. 2009 Sep 21;186(6):863-80. doi: 10.1083/jcb.200904111. J Cell Biol. 2009. PMID: 19797079 Free PMC article.
-
Genetic interactions of ribosome maturation factors Yvh1 and Mrt4 influence mRNA decay, glycogen accumulation, and the expression of early meiotic genes in Saccharomyces cerevisiae.J Biochem. 2011 Jul;150(1):103-11. doi: 10.1093/jb/mvr040. Epub 2011 Apr 6. J Biochem. 2011. PMID: 21474464
-
Mutational Analyses of the Cysteine-Rich Domain of Yvh1, a Protein Required for Translational Competency in Yeast.Biology (Basel). 2022 Aug 22;11(8):1246. doi: 10.3390/biology11081246. Biology (Basel). 2022. PMID: 36009873 Free PMC article.
-
Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast.Biochem J. 2017 Jan 15;474(2):195-214. doi: 10.1042/BCJ20160516. Biochem J. 2017. PMID: 28062837 Free PMC article. Review.
-
Insights into remodeling events during eukaryotic large ribosomal subunit assembly provided by high resolution cryo-EM structures.RNA Biol. 2017 Oct 3;14(10):1306-1313. doi: 10.1080/15476286.2017.1297914. Epub 2017 Mar 7. RNA Biol. 2017. PMID: 28267408 Free PMC article. Review.
Cited by
-
Ribosome-stalk biogenesis is coupled with recruitment of nuclear-export factor to the nascent 60S subunit.Nat Struct Mol Biol. 2016 Dec;23(12):1074-1082. doi: 10.1038/nsmb.3312. Epub 2016 Oct 24. Nat Struct Mol Biol. 2016. PMID: 27775710
-
Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1.Nat Struct Mol Biol. 2017 Mar;24(3):214-220. doi: 10.1038/nsmb.3364. Epub 2017 Jan 23. Nat Struct Mol Biol. 2017. PMID: 28112732 Free PMC article.
-
Zinc and the modulation of redox homeostasis.Free Radic Biol Med. 2012 Nov 1;53(9):1748-59. doi: 10.1016/j.freeradbiomed.2012.08.568. Epub 2012 Aug 25. Free Radic Biol Med. 2012. PMID: 22960578 Free PMC article. Review.
-
Ribosome biogenesis factors-from names to functions.EMBO J. 2023 Apr 3;42(7):e112699. doi: 10.15252/embj.2022112699. Epub 2023 Feb 10. EMBO J. 2023. PMID: 36762427 Free PMC article. Review.
-
Protein tyrosine phosphatase profiling studies during brown adipogenic differentiation of mouse primary brown preadipocytes.BMB Rep. 2013 Nov;46(11):539-43. doi: 10.5483/bmbrep.2013.46.11.058. BMB Rep. 2013. PMID: 24152912 Free PMC article.
References
-
- Ballesta J.P., Remacha M. 1996. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery.Prog. Nucleic Acid Res. Mol. Biol. 55:157–193 doi:10.1016/S0079-6603(08)60193-2 - DOI - PubMed
-
- Bécam A.M., Nasr F., Racki W.J., Zagulski M., Herbert C.J. 2001. Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae.Mol. Genet. Genomics. 266:454–462 doi:10.1007/s004380100548 - DOI - PubMed
-
- Beeser A.E., Cooper T.G. 2000. The dual-specificity protein phosphatase Yvh1p regulates sporulation, growth, and glycogen accumulation independently of catalytic activity in Saccharomyces cerevisiae via the cyclic AMP-dependent protein kinase cascade.J. Bacteriol. 182:3517–3528 doi:10.1128/JB.182.12.3517-3528.2000 - DOI - PMC - PubMed
-
- Berk V., Cate J.H. 2007. Insights into protein biosynthesis from structures of bacterial ribosomes.Curr. Opin. Struct. Biol. 17:302–309 doi:10.1016/j.sbi.2007.05.009 - DOI - PubMed
-
- Boguszewska A., Tchórzewski M., Dukowski P., Winiarczyk S., Grankowski N. 2002. Subcellular distribution of the acidic ribosomal P-proteins from Saccharomyces cerevisiae in various environmental conditions.Biol. Cell. 94:139–146 doi:10.1016/S0248-4900(02)01192-9 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

