Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5
- PMID: 19797429
- PMCID: PMC2775936
- DOI: 10.1210/me.2009-0242
Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5
Abstract
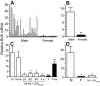
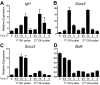
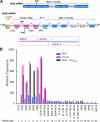
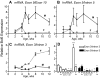
The transcriptional repressor Bcl6 is a male-specific rat liver gene product and one of 24 early GH-response genes encoding DNA-binding proteins. Presently, the sex specificity of Bcl6 was shown to emerge at puberty, when hepatic Bcl6 mRNA was induced in males and repressed in females by the female plasma GH profile. Hepatic Bcl6 mRNA was increased to near-normal male levels in hypophysectomized females and was extinguished in intact males given a continuous GH infusion (female-like GH pattern). Bcl6 was also repressed in adult male somatostatin-deficient mice, where plasma GH profiles are female like. Hepatic Bcl6 RNA was rapidly down-regulated by GH pulse treatment, both in hypophysectomized male rats and in primary rat hepatocytes. Bcl6 was substantially induced in female mice deficient in hepatic signal transducer and activator of transcription (STAT)5a/STAT5b, suggesting that these STAT transcriptional mediators of GH signaling repress Bcl6. Indeed, STAT5 was bound to Bcl6 STAT5-binding region-B, previously associated with Bcl6 repression, in both male and female liver chromatin. STAT5 also bound to Bcl6 region-A in male chromatin but only during a plasma GH pulse. Analysis of primary transcripts (heterogeneous nuclear RNA) across the Bcl6 gene revealed a novel mechanism of GH-dependent sex specificity, with two apparent blocks in Bcl6 transcription elongation seen in female liver and in continuous GH-treated male liver, one early in intron 4 and one in exon 5, which together reduced transcription beyond exon 5 more than 300-fold. Finally, Bcl6 was bound to a subset of STAT5-binding sites in male liver chromatin, including a Socs2 STAT5-binding site where Bcl6 binding increased substantially between plasma GH pulses, i.e. when STAT5 binding was low. Bcl6 and STAT5 binding are thus inversely coordinated by the endogenous pulses of pituitary GH release, suggesting this male-specific transcriptional repressor modulates hepatic GH signaling to select STAT5 target genes.
Figures



References
-
- Clodfelter KH, Holloway MG, Hodor P, Park SH, Ray WJ, Waxman DJ 2006 Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol 20:1333–1351 - PubMed
-
- Jansson JO, Edén S, Isaksson O 1985 Sexual dimorphism in the control of growth hormone secretion. Endocr Rev 6:128–150 - PubMed
-
- MacLeod JN, Pampori NA, Shapiro BH 1991 Sex differences in the ultradian pattern of plasma growth hormone concentrations in mice. J Endocrinol 131:395–399 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

