Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter
- PMID: 19797662
- PMCID: PMC4067766
- DOI: 10.1126/science.1175145
Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter
Abstract
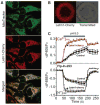
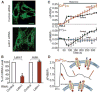
Mitochondria are integral components of cellular calcium (Ca2+) signaling. Calcium stimulates mitochondrial adenosine 5'-triphosphate production, but can also initiate apoptosis. In turn, cytoplasmic Ca2+ concentrations are regulated by mitochondria. Although several transporter and ion-channel mechanisms have been measured in mitochondria, the molecules that govern Ca2+ movement across the inner mitochondrial membrane are unknown. We searched for genes that regulate mitochondrial Ca2+ and H+ concentrations using a genome-wide Drosophila RNA interference (RNAi) screen. The mammalian homolog of one Drosophila gene identified in the screen, Letm1, was found to specifically mediate coupled Ca2+/H+ exchange. RNAi knockdown, overexpression, and liposome reconstitution of the purified Letm1 protein demonstrate that Letm1 is a mitochondrial Ca2+/H+ antiporter.
Figures


Comment in
-
Cell biology. A revolving door for calcium.Science. 2009 Oct 2;326(5949):57-8. doi: 10.1126/science.1180482. Science. 2009. PMID: 19797650 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

