Coexpression network based on natural variation in human gene expression reveals gene interactions and functions
- PMID: 19797678
- PMCID: PMC2775589
- DOI: 10.1101/gr.097600.109
Coexpression network based on natural variation in human gene expression reveals gene interactions and functions
Abstract
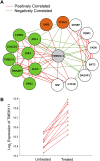
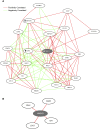
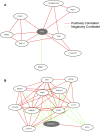
Genes interact in networks to orchestrate cellular processes. Analysis of these networks provides insights into gene interactions and functions. Here, we took advantage of normal variation in human gene expression to infer gene networks, which we constructed using correlations in expression levels of more than 8.5 million gene pairs in immortalized B cells from three independent samples. The resulting networks allowed us to identify biological processes and gene functions. Among the biological pathways, we found processes such as translation and glycolysis that co-occur in the same subnetworks. We predicted the functions of poorly characterized genes, including CHCHD2 and TMEM111, and provided experimental evidence that TMEM111 is part of the endoplasmic reticulum-associated secretory pathway. We also found that IFIH1, a susceptibility gene of type 1 diabetes, interacts with YES1, which plays a role in glucose transport. Furthermore, genes that predispose to the same diseases are clustered nonrandomly in the coexpression network, suggesting that networks can provide candidate genes that influence disease susceptibility. Therefore, our analysis of gene coexpression networks offers information on the role of human genes in normal and disease processes.
Figures


References
-
- Aggarwal BB, Henzel WJ, Moffat B, Kohr WJ, Harkins RN. Primary structure of human lymphotoxin derived from 1788 lymphoblastoid cell line. J Biol Chem. 1985;260:2334–2344. - PubMed
-
- Agrawal R, Imielinski T, Swami A. Mining association rules between sets of items in large databases; Proceedings of the 1993 ACM SIGMOD International Conference on Management of Data; Washington, D.C: ACM-SIGMOD; 1993. pp. 207–216.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
