Rapid identification of homologous recombinants and determination of gene copy number with reference/query pyrosequencing (RQPS)
- PMID: 19797679
- PMCID: PMC2775603
- DOI: 10.1101/gr.093856.109
Rapid identification of homologous recombinants and determination of gene copy number with reference/query pyrosequencing (RQPS)
Abstract
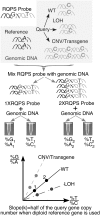
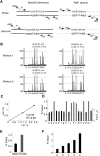
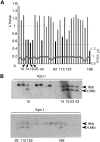
Manipulating the mouse genome is a widespread technology with important applications in many biological fields ranging from cancer research to developmental biology. Likewise, correlations between copy number variations (CNVs) and human diseases are emerging. We have developed the reference-query pyrosequencing (RQPS) method, which is based on quantitative pyrosequencing and uniquely designed probes containing single nucleotide variations (SNVs), to offer a simple and affordable genotyping solution capable of identifying homologous recombinants independent of the homology arm size, determining the micro-amplification status of endogenous human loci, and quantifying virus/transgene copy number in experimental or commercial species. In addition, we also present a simple pyrosequencing-based protocol that could be used for the enrichment of homologous recombinant embryonic stem (ES) cells.
Figures
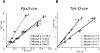
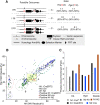
References
-
- Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. - PubMed
-
- Aten E, White SJ, Kalf ME, Vossen RH, Thygesen HH, Ruivenkamp CA, Kriek M, Breuning MH, den Dunnen JT. Methods to detect CNVs in the human genome. Cytogenet Genome Res. 2008;123:313–321. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
