A unified framework for detecting genetic association with multiple SNPs in a candidate gene or region: contrasting genotype scores and LD patterns between cases and controls
- PMID: 19797904
- PMCID: PMC2880731
- DOI: 10.1159/000243149
A unified framework for detecting genetic association with multiple SNPs in a candidate gene or region: contrasting genotype scores and LD patterns between cases and controls
Abstract
It is critical to develop and apply powerful statistical tests for genetic association studies due to typically weak associations with complex human diseases or phenotypes. For population-based case-control studies with unphased multilocus genotype data, most of the existing methods are based on comparing genotype scores, e.g. allele frequencies, between the case and control groups. Another class of approaches are motivated to contrast linkage disequilibrium (LD) patterns between the two groups. It is expected that no single test can be uniformly most powerful across all situations, and different tests may perform better under different scenarios. A recent effort has been devoted to combining the above two classes of approaches, which however has some potential drawbacks. Here we propose a general and simple framework to unify the above two classes of approaches: it is based on the simple idea to incorporate LD measurements, in addition to genotype scores, as covariates in a logistic regression model, from which various tests can be constructed by taking advantage of the nice properties of the score statistics for the logistic model. It also has an advantage in easily accommodating covariates and other study designs. We use simulated data to show that our proposed tests performed well across several scenarios. In particular, in contrast to either of the two classes of the tests that is only powerful in detecting only one, but not both, of the two types of the distributional differences between cases and controls, our proposed tests are sensitive to both.
Copyright 2009 S. Karger AG, Basel.
Figures
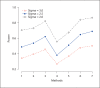
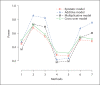
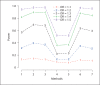
References
-
- Abecasis GR, Cookson WO. GOLD – graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. - PubMed
-
- Baksh MF, Balding DJ, Vyse TJ, Whittaker JC. A likelihood ratio approach to family-based association studies with covariates. Ann Hum Genet. 2006;70:131–139. - PubMed
-
- Cardon LR, Abecasis GR. Using haplotype blocks to map human complex trait loci. Trends Genet. 2003;19:135–140. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

