Failed gene conversion leads to extensive end processing and chromosomal rearrangements in fission yeast
- PMID: 19798055
- PMCID: PMC2776099
- DOI: 10.1038/emboj.2009.265
Failed gene conversion leads to extensive end processing and chromosomal rearrangements in fission yeast
Abstract
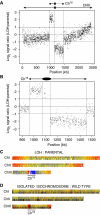
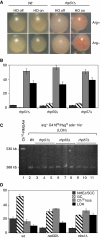
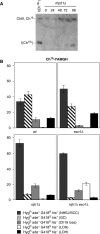
Loss of heterozygosity (LOH), a causal event in cancer and human genetic diseases, frequently encompasses multiple genetic loci and whole chromosome arms. However, the mechanisms by which such extensive LOH arises, and how it is suppressed in normal cells is poorly understood. We have developed a genetic system to investigate the mechanisms of DNA double-strand break (DSB)-induced extensive LOH, and its suppression, using a non-essential minichromosome, Ch(16), in fission yeast. We find extensive LOH to arise from a new break-induced mechanism of isochromosome formation. Our data support a model in which Rqh1 and Exo1-dependent end processing from an unrepaired DSB leads to removal of the broken chromosome arm and to break-induced replication of the intact arm from the centromere, a considerable distance from the initial lesion. This process also promotes genome-wide copy number variation. A genetic screen revealed Rhp51, Rhp55, Rhp57 and the MRN complex to suppress both isochromosome formation and chromosome loss, in accordance with these events resulting from extensive end processing associated with failed homologous recombination repair.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






Similar articles
-
Break-induced loss of heterozygosity in fission yeast: dual roles for homologous recombination in promoting translocations and preventing de novo telomere addition.Mol Cell Biol. 2007 Nov;27(21):7745-57. doi: 10.1128/MCB.00462-07. Epub 2007 Aug 27. Mol Cell Biol. 2007. PMID: 17724078 Free PMC article.
-
The DNA damage checkpoint pathway promotes extensive resection and nucleotide synthesis to facilitate homologous recombination repair and genome stability in fission yeast.Nucleic Acids Res. 2014 May;42(9):5644-56. doi: 10.1093/nar/gku190. Epub 2014 Mar 12. Nucleic Acids Res. 2014. PMID: 24623809 Free PMC article.
-
Mus81-Eme1-dependent and -independent crossovers form in mitotic cells during double-strand break repair in Schizosaccharomyces pombe.Mol Cell Biol. 2007 May;27(10):3828-38. doi: 10.1128/MCB.01596-06. Epub 2007 Mar 12. Mol Cell Biol. 2007. PMID: 17353272 Free PMC article.
-
Condensin-mediated chromosome organization in fission yeast.Curr Genet. 2016 Nov;62(4):739-743. doi: 10.1007/s00294-016-0601-7. Epub 2016 Apr 9. Curr Genet. 2016. PMID: 27061734 Free PMC article. Review.
-
Break-induced replication: what is it and what is it for?Cell Cycle. 2008 Apr 1;7(7):859-64. doi: 10.4161/cc.7.7.5613. Epub 2008 Jan 14. Cell Cycle. 2008. PMID: 18414031 Review.
Cited by
-
Gross Chromosomal Rearrangement at Centromeres.Biomolecules. 2023 Dec 24;14(1):28. doi: 10.3390/biom14010028. Biomolecules. 2023. PMID: 38254628 Free PMC article. Review.
-
The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA.Nat Commun. 2018 Oct 18;9(1):4340. doi: 10.1038/s41467-018-06545-y. Nat Commun. 2018. PMID: 30337534 Free PMC article. Review.
-
Heterochromatin suppresses gross chromosomal rearrangements at centromeres by repressing Tfs1/TFIIS-dependent transcription.Commun Biol. 2019 Jan 11;2:17. doi: 10.1038/s42003-018-0251-z. eCollection 2019. Commun Biol. 2019. PMID: 30652128 Free PMC article.
-
Fission yeast Srr1 and Skb1 promote isochromosome formation at the centromere.Commun Biol. 2023 May 26;6(1):551. doi: 10.1038/s42003-023-04925-9. Commun Biol. 2023. PMID: 37237082 Free PMC article.
-
Homologous recombination suppresses transgenerational DNA end resection and chromosomal instability in fission yeast.Nucleic Acids Res. 2023 Apr 24;51(7):3205-3222. doi: 10.1093/nar/gkad160. Nucleic Acids Res. 2023. PMID: 36951111 Free PMC article.
References
-
- Allshire R (2004) Centromere and kinetochore structure and function. In The molecular biology of Schizosaccharomyces pombe, Egel R (ed), Vol. 10, pp 150–169. Heidelberg: Springer-Verlag
-
- Andersen MK, Pedersen-Bjergaard J (2000) Increased frequency of dicentric chromosomes in therapy-related MDS and AML compared to de novo disease is significantly related to previous treatment with alkylating agents and suggests a specific susceptibility to chromosome breakage at the centromere. Leukemia 14: 105–111 - PubMed
-
- Atkin NB, Baker MC (1982) Specific chromosome change, i(12p), in testicular tumours? Lancet 2: 1349. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

