Interactome analyses identify ties of PrP and its mammalian paralogs to oligomannosidic N-glycans and endoplasmic reticulum-derived chaperones
- PMID: 19798432
- PMCID: PMC2749441
- DOI: 10.1371/journal.ppat.1000608
Interactome analyses identify ties of PrP and its mammalian paralogs to oligomannosidic N-glycans and endoplasmic reticulum-derived chaperones
Erratum in
- PLoS Pathog. 2009 Oct;5(10). doi: 10.1371/annotation/9eb11869-6acb-49b0-978e-abedc3cc545a. Won, Amy Hye [corrected to Jeon, Amy Hye Won] doi: 10.1371/annotation/9eb11869-6acb-49b0-978e-abedc3cc545a
Abstract
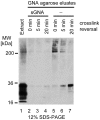
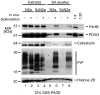
The physiological environment which hosts the conformational conversion of the cellular prion protein (PrP(C)) to disease-associated isoforms has remained enigmatic. A quantitative investigation of the PrP(C) interactome was conducted in a cell culture model permissive to prion replication. To facilitate recognition of relevant interactors, the study was extended to Doppel (Prnd) and Shadoo (Sprn), two mammalian PrP(C) paralogs. Interestingly, this work not only established a similar physiological environment for the three prion protein family members in neuroblastoma cells, but also suggested direct interactions amongst them. Furthermore, multiple interactions between PrP(C) and the neural cell adhesion molecule, the laminin receptor precursor, Na/K ATPases and protein disulfide isomerases (PDI) were confirmed, thereby reconciling previously separate findings. Subsequent validation experiments established that interactions of PrP(C) with PDIs may extend beyond the endoplasmic reticulum and may play a hitherto unrecognized role in the accumulation of PrP(Sc). A simple hypothesis is presented which accounts for the majority of interactions observed in uninfected cells and suggests that PrP(C) organizes its molecular environment on account of its ability to bind to adhesion molecules harboring immunoglobulin-like domains, which in turn recognize oligomannose-bearing membrane proteins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Taylor DR, Hooper NM. The prion protein and lipid rafts. Mol Membr Biol. 2006;23:89–99. - PubMed
-
- Watts JC, Westaway D. The prion protein family: diversity, rivalry, and dysfunction. Biochim Biophys Acta. 2007;1772:654–672. - PubMed
-
- Ben-Zaken O, Tzaban S, Tal Y, Horonchik L, Esko JD, et al. Cellular heparan sulfate participates in the metabolism of prions. J Biol Chem. 2003;278:40041–40049. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

