Solution structure of the complex of VEK-30 and plasminogen kringle 2
- PMID: 19800007
- PMCID: PMC2826548
- DOI: 10.1016/j.jsb.2009.09.011
Solution structure of the complex of VEK-30 and plasminogen kringle 2
Abstract
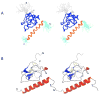
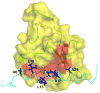
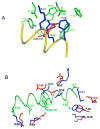
The solution structure of the complex containing the isolated kringle 2 domain of human plasminogen (K2(Pg)) and VEK-30, a 30-amino acid residue internal peptide from a streptococcal M-like plasminogen (Pg) binding protein (PAM), has been determined by multinuclear high-resolution NMR. Complete backbone and side-chain assignments were obtained from triple-resonance experiments, after which structure calculations were performed and ultimately refined by restrained molecular simulation in water. We find that, in contrast with the dimer of complexes observed in the asymmetric unit of the crystal, global correlation times and buoyant molecular weight determinations of the complex and its individual components showed the monomeric nature of all species in solution. The NMR-derived structure of K2(Pg) in complex with VEK-30 presents a folding pattern typical of other kringle domains, while bound VEK-30 forms an end-to-end alpha-helix (residues 6-27) in the complex. Most of the VEK-30/K2(Pg) interactions in solution occur between a single face of the alpha-helix of VEK-30 and the lysine binding site (LBS) of K2(Pg). The canonical LBS of K2(Pg), consisting of Asp54, Asp56, Trp60, Arg69, and Trp70 (kringle numbering), interacts with an internal pseudo-lysine of VEK-30, comprising side-chains of Arg17, His18, and Glu20. Site-specific mutagenesis analysis confirmed that the electrostatic field formed by the N-terminal anionic residues of the VEK-30 alpha-helix, viz., Asp7, and the non-conserved cationic residues of K2(Pg), viz., Lys43 and Arg55, play additional important roles in the docking of VEK-30 to K2(Pg). Structural analysis and kringle sequence alignments revealed several important features related to exosite binding that provide a structural rationale for the high specificity and affinity of VEK-30 for K2(Pg).
(c) 2009 Elsevier Inc. All rights reserved.
Figures


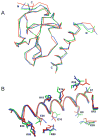
Similar articles
-
NMR backbone dynamics of VEK-30 bound to the human plasminogen kringle 2 domain.Biophys J. 2010 Jul 7;99(1):302-12. doi: 10.1016/j.bpj.2010.04.019. Biophys J. 2010. PMID: 20655859 Free PMC article.
-
Structure and binding determinants of the recombinant kringle-2 domain of human plasminogen to an internal peptide from a group A Streptococcal surface protein.J Mol Biol. 2001 May 11;308(4):705-19. doi: 10.1006/jmbi.2001.4646. J Mol Biol. 2001. PMID: 11350170
-
The lack of binding of VEK-30, an internal peptide from the group A streptococcal M-like protein, PAM, to murine plasminogen is due to two amino acid replacements in the plasminogen kringle-2 domain.J Biol Chem. 2008 Jan 18;283(3):1580-1587. doi: 10.1074/jbc.M705063200. Epub 2007 Nov 26. J Biol Chem. 2008. PMID: 18039665
-
Effects on human plasminogen conformation and activation rate caused by interaction with VEK-30, a peptide derived from the group A streptococcal M-like protein (PAM).Biochim Biophys Acta. 2010 Jun;1804(6):1342-9. doi: 10.1016/j.bbapap.2010.01.019. Epub 2010 Feb 10. Biochim Biophys Acta. 2010. PMID: 20152941 Free PMC article.
-
The kringle domains of human plasminogen.Ciba Found Symp. 1997;212:46-60; discussion 60-5. doi: 10.1002/9780470515457.ch4. Ciba Found Symp. 1997. PMID: 9524763 Review.
Cited by
-
Conformationally organized lysine isosteres in Streptococcus pyogenes M protein mediate direct high-affinity binding to human plasminogen.J Biol Chem. 2017 Sep 8;292(36):15016-15027. doi: 10.1074/jbc.M117.794198. Epub 2017 Jul 19. J Biol Chem. 2017. PMID: 28724633 Free PMC article.
-
NMR backbone dynamics of VEK-30 bound to the human plasminogen kringle 2 domain.Biophys J. 2010 Jul 7;99(1):302-12. doi: 10.1016/j.bpj.2010.04.019. Biophys J. 2010. PMID: 20655859 Free PMC article.
-
Direct Host Plasminogen Binding to Bacterial Surface M-protein in Pattern D Strains of Streptococcus pyogenes Is Required for Activation by Its Natural Coinherited SK2b Protein.J Biol Chem. 2015 Jul 24;290(30):18833-42. doi: 10.1074/jbc.M115.655365. Epub 2015 Jun 12. J Biol Chem. 2015. PMID: 26070561 Free PMC article.
-
Binding of the kringle-2 domain of human plasminogen to streptococcal PAM-type M-protein causes dissociation of PAM dimers.Microbiologyopen. 2021 Nov;10(6):e1252. doi: 10.1002/mbo3.1252. Microbiologyopen. 2021. PMID: 34964287 Free PMC article.
-
Affinity maturation, humanization, and co-crystallization of a rabbit anti-human ROR2 monoclonal antibody for therapeutic applications.J Biol Chem. 2020 May 1;295(18):5995-6006. doi: 10.1074/jbc.RA120.012791. Epub 2020 Mar 19. J Biol Chem. 2020. PMID: 32193207 Free PMC article.
References
-
- Sottrup-Jensen L, Claeys H, Zajdel M, Petersen TE, Magnusson S. The primary structure of human plasminogen: isolation of two lysine-binding fragments and one “mini” plasminogen (MW, 38000) by elastase-catalyzed-specific limited proteolysis. Prog Chem Fibrinolysis Thrombolysis. 1978;3:191–209.
-
- Menhart N, Sehl LC, Kelley RF, Castellino FJ. Construction, expression and purification of recombinant kringle 1 of human plasminogen and analysis of its interaction with ω-amino acids. Biochemistry. 1991;30:1948–1957. - PubMed
-
- Hoover GJ, Menhart N, Martin A, Warder S, Castellino FJ. Amino acids of the recombinant kringle 1 domain of human plasminogen that stabilize its interaction with ω-amino acids. Biochemistry. 1993;32:10936–10943. - PubMed
-
- McCance SG, Menhart N, Castellino FJ. Amino acid residues of the kringle-4 and kringle-5 domains of human plasminogen that stabilize their interactions with omega-amino acid ligands. J Biol Chem. 1994;269:32405–32410. - PubMed
-
- Nilsen SL, Prorok M, Castellino FJ. Enhancement through mutagenesis of the binding of the isolated kringle 2 domain of human plasminogen to omega-amino acid ligands and to an internal sequence of a Streptococcal surface protein. J Biol Chem. 1999;274:22380–22386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

