A lymphotoxin-driven pathway to hepatocellular carcinoma
- PMID: 19800575
- PMCID: PMC4422166
- DOI: 10.1016/j.ccr.2009.08.021
A lymphotoxin-driven pathway to hepatocellular carcinoma
Erratum in
- Cancer Cell. 2009 Nov 6;16(5):447
-
A Lymphotoxin-Driven Pathway to Hepatocellular Carcinoma.Cancer Cell. 2025 Oct 13;43(10):1968-1972. doi: 10.1016/j.ccell.2025.08.005. Epub 2025 Sep 28. Cancer Cell. 2025. PMID: 41016394 No abstract available.
Abstract
Hepatitis B and C viruses (HBV and HCV) cause chronic hepatitis and hepatocellular carcinoma (HCC) by poorly understood mechanisms. We show that cytokines lymphotoxin (LT) alpha and beta and their receptor (LTbetaR) are upregulated in HBV- or HCV-induced hepatitis and HCC. Liver-specific LTalphabeta expression in mice induces liver inflammation and HCC, causally linking hepatic LT overexpression to hepatitis and HCC. Development of HCC, composed in part of A6(+) oval cells, depends on lymphocytes and IKappa B kinase beta expressed by hepatocytes but is independent of TNFR1. In vivo LTbetaR stimulation implicates hepatocytes as the major LT-responsive liver cells, and LTbetaR inhibition in LTalphabeta-transgenic mice with hepatitis suppresses HCC formation. Thus, sustained LT signaling represents a pathway involved in hepatitis-induced HCC.
Figures

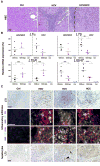


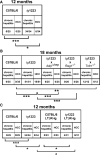



Comment in
-
Lymphotoxins: new targets for hepatocellular carcinoma.Cancer Cell. 2009 Oct 6;16(4):272-3. doi: 10.1016/j.ccr.2009.09.012. Cancer Cell. 2009. PMID: 19800570 Free PMC article.
References
-
- Akhurst B, Matthews V, Husk K, Smyth MJ, Abraham LJ, Yeoh GC. Differential lymphotoxin-beta and interferon gamma signaling during mouse liver regeneration induced by chronic and acute injury. Hepatology. 2005;41:327–335. - PubMed
-
- An MM, Fan KX, Cao YB, Shen H, Zhang JD, Lu L, Gao PH, Jiang YY. Lymphtoxin beta receptor-Ig protects from T-cell-mediated liver injury in mice through blocking LIGHT/HVEM signaling. Biol Pharm Bull. 2006;29:2025–2030. - PubMed
-
- Bachert C, Fimmel C, Linstedt AD. Endosomal trafficking and proprotein convertase cleavage of cis Golgi protein GP73 produces marker for hepatocellular carcinoma. Traffic. 2007;8:1415–1423. - PubMed
-
- Baffet G, Braciak TA, Fletcher RG, Gauldie J, Fey GH, Northemann W. Autocrine activity of interleukin 6 secreted by hepatocarcinoma cell lines. Mol Biol Med. 1991;8:141–156. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

