Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model
- PMID: 19800577
- PMCID: PMC2829755
- DOI: 10.1016/j.ccr.2009.08.015
Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model
Abstract
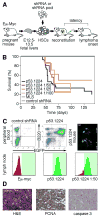
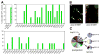
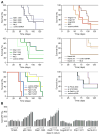
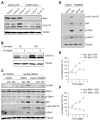
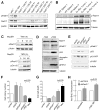
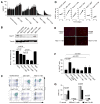
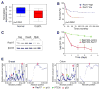
Short hairpin RNAs (shRNAs) capable of stably suppressing gene function by RNA interference (RNAi) can mimic tumor-suppressor-gene loss in mice. By selecting for shRNAs capable of accelerating lymphomagenesis in a well-characterized mouse lymphoma model, we identified over ten candidate tumor suppressors, including Sfrp1, Numb, Mek1, and Angiopoietin 2. Several components of the DNA damage response machinery were also identified, including Rad17, which acts as a haploinsufficient tumor suppressor that responds to oncogenic stress and whose loss is associated with poor prognosis in human patients. Our results emphasize the utility of in vivo RNAi screens, identify and validate a diverse set of tumor suppressors, and have therapeutic implications.
Figures
References
-
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
-
- Bao S, Tibbetts RS, Brumbaugh KM, Fang Y, Richardson DA, Ali A, Chen SM, Abraham RT, Wang XF. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature. 2001;411:969–974. - PubMed
-
- Beretta GL, Gatti L, Cesare MD, Corna E, Tinelli S, Carenini N, Zunino F, Perego P. The human homolog of fission yeast Rad17 is implicated in tumor growth. Cancer Lett 2008 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

