Carbohydrate recognition by the mannose-6-phosphate receptors
- PMID: 19801188
- PMCID: PMC2771201
- DOI: 10.1016/j.sbi.2009.09.002
Carbohydrate recognition by the mannose-6-phosphate receptors
Abstract
The two P-type lectins, the 46kDa cation-dependent mannose-6-phosphate (Man-6-P) receptor (CD-MPR), and the 300kDa cation-independent Man-6-P receptor (CI-MPR), are the founding members of the growing family of mannose-6-phosphate receptor homology (MRH) proteins. A major cellular function of the MPRs is to transport Man-6-P-containing acid hydrolases from the Golgi to endosomal/lysosomal compartments. Recent advances in the structural analyses of both CD-MPR and CI-MPR have revealed the structural basis for phosphomannosyl recognition by these receptors and provided insights into how the receptors load and unload their cargo. A surprising finding is that the CD-MPR is dynamic, with at least two stable quaternary states, the open (ligand-bound) and closed (ligand-free) conformations, similar to those of hemoglobin. Ligand binding stabilizes the open conformation; changes in the pH of the environment at the cell surface and in endosomal compartments weaken the ligand-receptor interaction and/or weaken the electrostatic interactions at the subunit interface, resulting in the closed conformation.
Figures

 ). The dimeric CD-MPR is depicted as two pink balls and the CI-MPR is shown as 15 repeating balls. The three Man-6-P binding domains of the CI-MPR are depicted as pink balls.
). The dimeric CD-MPR is depicted as two pink balls and the CI-MPR is shown as 15 repeating balls. The three Man-6-P binding domains of the CI-MPR are depicted as pink balls.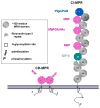



References
-
- Gelfman CM, Vogel P, Issa TM, Turner CA, Lee WS, Kornfeld S, Rice DS. Mice lacking alpha/beta subunits of GlcNAc-1-phosphotransferase exhibit growth retardation, retinal degeneration, and secretory cell lesions. Invest Ophthalmol Vis Sci. 2007;48:5221–5228. GlcNac-1 phosphotransferase catalyzes the first step of the two step process in the acquisition of mannose 6-phosphate on N-linked oligosaccharides of lysosomal enzymes. Mice deficient in the phosphotransferase gene exhibit severe retinal degeneration in addition to the features observed in patients with mucolipidosis II. Authors suggest a connection between retinal diseases and lysosomal storage disorders via GlcNac-phosphotransferase. - PubMed
-
- Lee WS, Payne BJ, Gelfman CM, Vogel P, Kornfeld S. Murine UDP-GlcNAc:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase lacking the gamma-subunit retains substantial activity toward acid hydrolases. J Biol Chem. 2007;282:27198–27203. Together with Ref. 1, this article characterizes the phosphotransferase. The paper shows that the α/βsubunits of the enzyme, in addition to their catalytic function, have some ability to recognize acid hydrolases as specific substrates. - PubMed
-
- Varki A, Sherman W, Kornfeld S. Demonstration of the enzymatic mechanisms of alpha-N-acetyl-D-glucosamine-1-phosphodiester N-acetylglucosaminidase (formerly called alpha-N-acetylglucosaminylphosphodiesterase) and lysosomal alpha-N-acetylglucosaminidase. Archives of Biochemistry & Biophysics. 1983;222:145–149. - PubMed
-
- Le Borgne R, Hoflack B. Protein transport from the secretory to the endocytic pathway in mammalian cells. Biochim Biophys Acta. 1998;1404:195–209. - PubMed
-
- Mullins C, Bonifacino JS. The molecular machinery for lysosome biogenesis. Bioessays. 2001;23:333–343. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

