A family of transcriptional antitermination factors necessary for synthesis of the capsular polysaccharides of Bacteroides fragilis
- PMID: 19801412
- PMCID: PMC2786560
- DOI: 10.1128/JB.00500-09
A family of transcriptional antitermination factors necessary for synthesis of the capsular polysaccharides of Bacteroides fragilis
Abstract
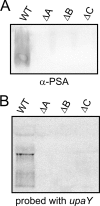
A single strain of Bacteroides fragilis synthesizes eight distinct capsular polysaccharides, designated PSA to PSH. These polysaccharides are synthesized by-products encoded by eight separate polysaccharide biosynthesis loci. The genetic architecture of each of these eight loci is similar, including the fact that the first gene of each locus is a paralog of the first gene of each of the other PS loci. These proteins are designated the UpxY family, where x is replaced by a to h, depending upon the polysaccharide locus from which it is produced. Mutational analysis of three separate upxY genes demonstrated that they are necessary and specific for transcription of their respective polysaccharide biosynthesis operon and that they function in trans. Transcriptional reporter constructs, reverse transcriptase PCR, and deletion analysis demonstrated that the UpxYs do not affect initiation of transcription, but rather prevent premature transcriptional termination within the 5' untranslated region between the promoter and the upxY gene. The UpxYs have conserved motifs that are present in NusG and NusG-like proteins. Mutation of two conserved residues within the conserved KOW motif abrogated UpaY activity, further confirming that these proteins belong to the NusG-like (NusG(SP)) family. Alignment of highly similar UpxYs led to the identification of a small region of these proteins predicted to confer specificity for their respective loci. Construction of an upaY-upeY hybrid that produced a protein in which a 17-amino-acid segment of UpaY was changed to that of UpeY altered UpaY's specificity, as it was now able to function in transcriptional antitermination of the PSE biosynthesis operon.
Figures



References
-
- Artsimovitch, I., and R. Landick. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109:193-203. - PubMed
-
- Bailey, M. J., C. Hughes, and V. Koronakis. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26:845-851. - PubMed
-
- Baumann, H., A. O. Tzianabos, J. R. Brisson, D. L. Kasper, and H. J. Jennings. 1992. Structural elucidation of two capsular polysaccharides from one strain of Bacteroides fragilis using high-resolution NMR spectroscopy. Biochemistry 31:4081-4089. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

