Molecular regulation of arabinan and L-arabinose metabolism in Hypocrea jecorina (Trichoderma reesei)
- PMID: 19801419
- PMCID: PMC2794218
- DOI: 10.1128/EC.00162-09
Molecular regulation of arabinan and L-arabinose metabolism in Hypocrea jecorina (Trichoderma reesei)
Abstract
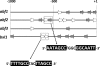
Hypocrea jecorina (anamorph: Trichoderma reesei) can grow on plant arabinans by the aid of secreted arabinan-degrading enzymes. This growth on arabinan and its degradation product L-arabinose requires the operation of the aldose reductase XYL1 and the L-arabinitol dehydrogenase LAD1. Growth on arabinan and L-arabinose is also severely affected in a strain deficient in the general cellulase and hemicellulase regulator XYR1, but this impairment can be overcome by constitutive expression of the xyl1 encoding the aldose reductase. An inspection of the genome of H. jecorina reveals four genes capable of degrading arabinan, i.e., the alpha-L-arabinofuranosidase encoding genes abf1, abf2, and abf3 and also bxl1, which encodes a beta-xylosidase with a separate alpha-L-arabinofuranosidase domain and activity but no endo-arabinanase. Transcriptional analysis reveals that in the parent strain QM9414 the expression of all of these genes is induced by L-arabinose and to a lesser extent by L-arabinitol and absent on D-glucose. Induction by L-arabinitol, however, is strongly enhanced in a Deltalad1 strain lacking L-arabinitol dehydrogenase activity and severely impaired in an aldose reductase (Deltaxyl1) strain, suggesting a cross talk between L-arabinitol and the aldose reductase XYL1 in an alpha-L-arabinofuranosidase gene expression. Strains bearing a knockout in the cellulase regulator xyr1 do not show any induction of abf2 and bxl1, and this phenotype cannot be reverted by constitutive expression of xyl1. The loss of function of xyr1 has also a slight effect on the expression of abf1 and abf3. We conclude that the expression of the four alpha-L-arabinofuranosidases of H. jecorina for growth on arabinan requires an early pathway intermediate (L-arabinitol or L-arabinose), the first enzyme of the pathway XYL1, and in the case of abf2 and bxl1 also the function of the cellulase regulator XYR1.
Figures

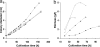




References
-
- Alexiou, P., K. Pegklidou, M. Chatzopoulou, I. Nicolaou, and V. J. Demopoulos. 2009. Aldose reductase enzyme and its implication to major health problems of the 21(st) century. Curr. Med. Chem. 16:734-752. - PubMed
-
- Ausubel, F. M., B. Roger, R. E. Kingston, D. D. Moore, J. G. Seidman, J. Smith, and K. Struhl. 2006. Current protocols in molecular biology. Greene Publishing Associates/Wiley Interscience, New York, NY.
-
- Bhardwaj, A., and M. F. Wilkinson. 2005. A metabolic enzyme doing double duty as a transcription factor. Bioessays 27:467-471. - PubMed
-
- Carpita, N. C. 1996. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Physiol. Plant Mol. Biol. 47:445-476. - PubMed
-
- Cho, Y. H., S. D. Yoo, and J. Sheen, J. 2006. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127:579-589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

