Structure-based mechanism of ADP-ribosylation by sirtuins
- PMID: 19801667
- PMCID: PMC2785207
- DOI: 10.1074/jbc.M109.024521
Structure-based mechanism of ADP-ribosylation by sirtuins
Abstract
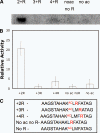
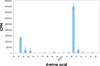
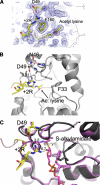
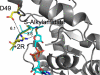
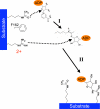
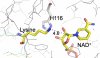
Sirtuins comprise a family of enzymes found in all organisms, where they play a role in diverse processes including transcriptional silencing, aging, regulation of transcription, and metabolism. The predominant reaction catalyzed by these enzymes is NAD(+)-dependent lysine deacetylation, although some sirtuins exhibit a weaker ADP-ribosyltransferase activity. Although the Sir2 deacetylation mechanism is well established, much less is known about the Sir2 ADP-ribosylation reaction. We have studied the ADP-ribosylation activity of a bacterial sirtuin, Sir2Tm, and show that acetylated peptides containing arginine or lysine 2 residues C-terminal to the acetyl lysine, the +2 position, are preferentially ADP-ribosylated at the +2 residue. A structure of Sir2Tm bound to the acetylated +2 arginine peptide shows how this arginine could enter the active site and react with a deacetylation reaction intermediate to yield an ADP-ribosylated peptide. The new biochemical and structural studies presented here provide mechanistic insights into the Sir2 ADP-ribosylation reaction and will aid in identifying substrates of this reaction.
Figures
References
-
- Frye R. A. (2000) Biochem. Biophys. Res. Commun. 273, 793–798 - PubMed
-
- Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. (2004) Cell 116, 551–563 - PubMed
-
- Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Cell 107, 137–148 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

