The structure of the N-terminus of kindlin-1: a domain important for alphaiibbeta3 integrin activation
- PMID: 19804783
- PMCID: PMC2963925
- DOI: 10.1016/j.jmb.2009.09.061
The structure of the N-terminus of kindlin-1: a domain important for alphaiibbeta3 integrin activation
Abstract
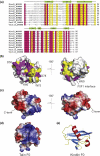
The integrin family of heterodimeric cell adhesion molecules exists in both low- and high-affinity states, and integrin activation requires binding of the talin FERM (four-point-one, ezrin, radixin, moesin) domain to membrane-proximal sequences in the beta-integrin cytoplasmic domain. However, it has recently become apparent that the kindlin family of FERM domain proteins is also essential for talin-induced integrin activation. FERM domains are typically composed of F1, F2, and F3 domains, but the talin FERM domain is atypical in that it contains a large insert in F1 and is preceded by a previously unrecognized domain, F0. Initial sequence alignments showed that the kindlin FERM domain was most similar to the talin FERM domain, but the homology appeared to be restricted to the F2 and F3 domains. Based on a detailed characterization of the talin FERM domain, we have reinvestigated the sequence relationship with kindlins and now show that kindlins do indeed contain the same domain structure as the talin FERM domain. However, the kindlin F1 domain contains an even larger insert than that in talin F1 that disrupts the sequence alignment. The insert, which varies in length between different kindlins, is not conserved and, as in talin, is largely unstructured. We have determined the structure of the kindlin-1 F0 domain by NMR, which shows that it adopts the same ubiquitin-like fold as the talin F0 and F1 domains. Comparison of the kindlin-1 and talin F0 domains identifies the probable interface with the kindlin-1 F1 domain. Potential sites of interaction of kindlin F0 with other proteins are discussed, including sites that differ between kindlin-1, kindlin-2, and kindlin-3. We also demonstrate that F0 is required for the ability of kindlin-1 to support talin-induced alphaIIbbeta3 integrin activation and for the localization of kindlin-1 to focal adhesions.
Figures




References
-
- Aszodi A., Legate K.R., Nakchbandi I., Fassler R. What mouse mutants teach us about extracellular matrix function. Annu. Rev. Cell Dev. Biol. 2006;22:591–621. - PubMed
-
- Legate K.R., Fassler R. Mechanisms that regulate adaptor binding to β-integrin cytoplasmic tails. J. Cell Sci. 2009;122:187–198. - PubMed
-
- Legate K.R., Montanez E., Kudlacek O., Fassler R. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat. Rev., Mol. Cell Biol. 2006;7:20–31. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

