Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA
- PMID: 19804849
- PMCID: PMC2756550
- DOI: 10.1016/j.ajhg.2009.09.007
Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA
Abstract
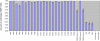
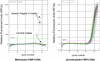
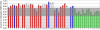
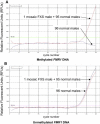
Fragile X syndrome (FXS) results from a CGG-repeat expansion that triggers hypermethylation and silencing of the FMR1 gene. FXS is referred to as the most common form of inherited intellectual disability, yet its true incidence has never been measured directly by large population screening. Here, we developed an inexpensive and high-throughput assay to quantitatively assess FMR1 methylation in DNA isolated from the dried blood spots of 36,124 deidentified newborn males. This assay displays 100% specificity and 100% sensitivity for detecting FMR1 methylation, successfully distinguishing normal males from males with full-mutation FXS. Furthermore, the assay can detect excess FMR1 methylation in 82% of females with full mutations, although the methylation did not correlate with intellectual disability. With amelogenin PCR used for detecting the presence of a Y chromosome, this assay can also detect males with Klinefelter syndrome (KS) (47, XXY). We identified 64 males with FMR1 methylation and, after confirmatory testing, found seven to have full-mutation FXS and 57 to have KS. Because the precise incidence of KS is known, we used our observed KS incidence as a sentinel to assess ascertainment quality and showed that our KS incidence of 1 in 633 newborn males was not significantly different from the literature incidence of 1 in 576 (p = 0.79). The seven FXS males revealed an FXS incidence in males of 1 in 5161 (95% confidence interval of 1 in 10,653-1 in 2500), consistent with some earlier indirect estimates. Given the trials now underway for possible FXS treatments, this method could be used in newborn or infant screening as a way of ensuring early interventions for FXS.
Figures

References
-
- Pieretti M., Zhang F.P., Fu Y.H., Warren S.T., Oostra B.A., Caskey C.T., Nelson D.L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. - PubMed
-
- Fu Y.H., Kuhl D.P., Pizzuti A., Pieretti M., Sutcliffe J.S., Richards S., Verkerk A.J., Holden J.J., Fenwick R.G., Warren S.T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. - PubMed
-
- Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. - PubMed
-
- Sutcliffe J.S., Nelson D.L., Zhang F., Pieretti M., Caskey C.T., Saxe D., Warren S.T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1992;1:397–400. - PubMed
-
- Eberhart D.E., Warren S.T. Nuclease sensitivity of permeabilized cells confirms altered chromatin formation at the fragile X locus. Somat. Cell Mol. Genet. 1996;22:435–441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

