Quantum-mechanics-derived 13Calpha chemical shift server (CheShift) for protein structure validation
- PMID: 19805131
- PMCID: PMC2761357
- DOI: 10.1073/pnas.0908833106
Quantum-mechanics-derived 13Calpha chemical shift server (CheShift) for protein structure validation
Abstract
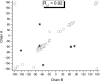
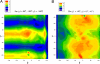
A server (CheShift) has been developed to predict (13)C(alpha) chemical shifts of protein structures. It is based on the generation of 696,916 conformations as a function of the phi, psi, omega, chi1 and chi2 torsional angles for all 20 naturally occurring amino acids. Their (13)C(alpha) chemical shifts were computed at the DFT level of theory with a small basis set and extrapolated, with an empirically-determined linear regression formula, to reproduce the values obtained with a larger basis set. Analysis of the accuracy and sensitivity of the CheShift predictions, in terms of both the correlation coefficient R and the conformational-averaged rmsd between the observed and predicted (13)C(alpha) chemical shifts, was carried out for 3 sets of conformations: (i) 36 x-ray-derived protein structures solved at 2.3 A or better resolution, for which sets of (13)C(alpha) chemical shifts were available; (ii) 15 pairs of x-ray and NMR-derived sets of protein conformations; and (iii) a set of decoys for 3 proteins showing an rmsd with respect to the x-ray structure from which they were derived of up to 3 A. Comparative analysis carried out with 4 popular servers, namely SHIFTS, SHIFTX, SPARTA, and PROSHIFT, for these 3 sets of conformations demonstrated that CheShift is the most sensitive server with which to detect subtle differences between protein models and, hence, to validate protein structures determined by either x-ray or NMR methods, if the observed (13)C(alpha) chemical shifts are available. CheShift is available as a web server.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. - PubMed
-
- Williamson MP, Craven CJ. Automated protein structure calculation from NMR data. J Biomol NMR. 2009;43:131–143. - PubMed
-
- Vriend G. WHAT IF: A molecular modeling and drug design program. J Mol Graphics. 1990;8:52–56. - PubMed
-
- Lüthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

