Nod2 is required for the regulation of commensal microbiota in the intestine
- PMID: 19805227
- PMCID: PMC2747201
- DOI: 10.1073/pnas.0907722106
Nod2 is required for the regulation of commensal microbiota in the intestine
Abstract
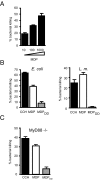
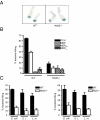
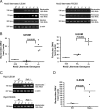
Mutations in the Nod2 gene are among the strongest genetic risk factors in the pathogenesis of ileal Crohn's disease, but the exact contributions of Nod2 to intestinal mucosal homeostasis are not understood. Here we show that Nod2 plays an essential role in controlling commensal bacterial flora in the intestine. Analysis of intestinal bacteria from the terminal ilea of Nod2-deficient mice showed that they harbor an increased load of commensal resident bacteria. Furthermore, Nod2-deficient mice had a diminished ability to prevent intestinal colonization of pathogenic bacteria. In vitro, intestinal crypts isolated from terminal ilea of Nod2-deficient mice were unable to kill bacteria effectively, suggesting an important role of Nod2 signaling in crypt function. Interestingly, the expression of Nod2 is dependent on the presence of commensal bacteria, because mice re-derived into germ-free conditions expressed significantly less Nod2 in their terminal ilea, and complementation of commensal bacteria into germ-free mice induced Nod2 expression. Therefore, Nod2 and intestinal commensal bacterial flora maintain a balance by regulating each other through a feedback mechanism. Dysfunction of Nod2 results in a break-down of this homeostasis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Recognition of commensal microbes: if innate responses are NOD in balance.Expert Rev Clin Immunol. 2010 Mar;6(2):205-10. doi: 10.1586/eci.10.1. Expert Rev Clin Immunol. 2010. PMID: 20402383 No abstract available.
References
-
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. - PubMed
-
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. - PubMed
-
- Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. - PubMed
-
- Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

