Organization of cytochrome P450 enzymes involved in sex steroid synthesis: PROTEIN-PROTEIN INTERACTIONS IN LIPID MEMBRANES
- PMID: 19805543
- PMCID: PMC2785165
- DOI: 10.1074/jbc.M109.006064
Organization of cytochrome P450 enzymes involved in sex steroid synthesis: PROTEIN-PROTEIN INTERACTIONS IN LIPID MEMBRANES
Abstract
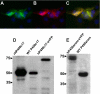
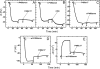
Mounting evidence underscores the importance of protein-protein interactions in the functional regulation of drug-metabolizing P450s, but few studies have been conducted in membrane environments, and none have examined P450s catalyzing sex steroid synthesis. Here we report specific protein-protein interactions for full-length, human, wild type steroidogenic cytochrome P450 (P450, CYP) enzymes: 17alpha-hydroxylase/17,20-lyase (P450c17, CYP17) and aromatase (P450arom, CYP19), as well as their electron donor NADPH-cytochrome P450 oxidoreductase (CPR). Fluorescence resonance energy transfer (FRET)(3) in live cells, coupled with quartz crystal microbalance (QCM), and atomic force microscopy (AFM) studies on phosphatidyl choline +/- cholesterol (mammalian) biomimetic membranes were used to investigate steroidogenic P450 interactions. The FRET results in living cells demonstrated that both P450c17 and P450arom homodimerize but do not heterodimerize, although they each heterodimerize with CPR. The lack of heteroassociation between P450c17 and P450arom was confirmed by QCM, wherein neither enzyme bound a membrane saturated with the other. In contrast, the CPR bound readily to either P450c17- or P450arom-saturated surfaces. Interestingly, N-terminally modified P450arom was stably incorporated and gave similar results to the wild type, although saturation was achieved with much less protein, suggesting that the putative transmembrane domain is not required for membrane association but for orientation. In fact, all of the proteins were remarkably stable in the membrane, such that high resolution AFM images were obtained, further supporting the formation of P450c17, P450arom, and CPR homodimers and oligomers in lipid bilayers. This unique combination of in vivo and in vitro studies has provided strong evidence for homodimerization and perhaps some higher order interactions for both P450c17 and P450arom.
Figures



References
-
- Conley A., Hinshelwood M. (2001) Reproduction 121, 685–695 - PubMed
-
- Hillier S. G., Whitelaw P. F., Smyth C. D. (1994) Mol. Cell. Endocrinol. 100, 51–54 - PubMed
-
- Conley A. J., Mason J. I. (1990) Baillieres Clin. Endo. Metab. 4, 249–272 - PubMed
-
- Backes W. L., Kelley R. W. (2003) Pharmacol. Ther. 98, 221–233 - PubMed
-
- Szczesna-Skorupa E., Mallah B., Kemper B. (2003) J. Biol. Chem. 278, 31269–31276 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

