Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation
- PMID: 19805909
- PMCID: PMC2769176
- DOI: 10.1172/JCI39199
Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation
Abstract
The development of highly active antiretroviral therapy (HAART) to treat individuals infected with HIV-1 has dramatically improved patient outcomes, but HAART still fails to cure the infection. The latent viral reservoir in resting CD4+ T cells is a major barrier to virus eradication. Elimination of this reservoir requires reactivation of the latent virus. However, strategies for reactivating HIV-1 through nonspecific T cell activation have clinically unacceptable toxicities. We describe here the development of what we believe to be a novel in vitro model of HIV-1 latency that we used to search for compounds that can reverse latency. Human primary CD4+ T cells were transduced with the prosurvival molecule Bcl-2, and the resulting cells were shown to recapitulate the quiescent state of resting CD4+ T cells in vivo. Using this model system, we screened small-molecule libraries and identified a compound that reactivated latent HIV-1 without inducing global T cell activation, 5-hydroxynaphthalene-1,4-dione (5HN). Unlike previously described latency-reversing agents, 5HN activated latent HIV-1 through ROS and NF-kappaB without affecting nuclear factor of activated T cells (NFAT) and PKC, demonstrating that TCR pathways can be dissected and utilized to purge latent virus. Our study expands the number of classes of latency-reversing therapeutics and demonstrates the utility of this in vitro model for finding strategies to eradicate HIV-1 infection.
Figures

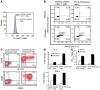

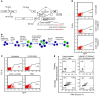
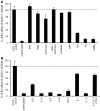
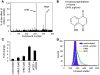


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

