Draft genome sequence of the Daphnia pathogen Octosporea bayeri: insights into the gene content of a large microsporidian genome and a model for host-parasite interactions
- PMID: 19807911
- PMCID: PMC2784321
- DOI: 10.1186/gb-2009-10-10-r106
Draft genome sequence of the Daphnia pathogen Octosporea bayeri: insights into the gene content of a large microsporidian genome and a model for host-parasite interactions
Abstract
Background: The highly compacted 2.9-Mb genome of Encephalitozoon cuniculi placed the microsporidia in the spotlight, encoding a mere 2,000 proteins and a highly reduced suite of biochemical pathways. This extreme level of reduction is not universal across the microsporidia, with genomes known to vary up to sixfold in size, suggesting that some genomes may harbor a gene content that is not as reduced as that of Enc. cuniculi. In this study, we present an in-depth survey of the large genome of Octosporea bayeri, a pathogen of Daphnia magna, with an estimated genome size of 24 Mb, in order to shed light on the organization and content of a large microsporidian genome.
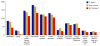
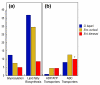
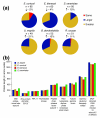
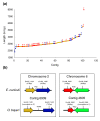
Results: Using Illumina sequencing, 898 Mb of O. bayeri genome sequence was generated, resulting in 13.3 Mb of unique sequence. We annotated a total of 2,174 genes, of which 893 encodes proteins with assigned function. The gene density of the O. bayeri genome is very low on average, but also highly uneven, so gene-dense regions also occur. The data presented here suggest that the O. bayeri proteome is well represented in this analysis and is more complex that that of Enc. cuniculi. Functional annotation of O. bayeri proteins suggests that this species might be less biochemically dependent on its host for its metabolism than its more reduced relatives.
Conclusions: The combination of the data presented here, together with the imminent annotated genome of Daphnia magna, will provide a wealth of genetic and genomic tools to study host-parasite interactions in an interesting model for pathogenesis.
Figures

Similar articles
-
The Ordospora colligata genome: Evolution of extreme reduction in microsporidia and host-to-parasite horizontal gene transfer.mBio. 2015 Jan 13;6(1):e02400-14. doi: 10.1128/mBio.02400-14. mBio. 2015. PMID: 25587016 Free PMC article.
-
Phylogenetic position of Octosporea muscaedomesticae (Microsporidia) and its relationship to Octosporea bayeri based on small subunit rDNA analysis.J Invertebr Pathol. 2010 Nov;105(3):366-70. doi: 10.1016/j.jip.2010.07.007. Epub 2010 Aug 3. J Invertebr Pathol. 2010. PMID: 20688077
-
Octosporea bayeri: fumidil B inhibits vertical transmission in Daphnia magna.Exp Parasitol. 2005 Jan;109(1):58-61. doi: 10.1016/j.exppara.2004.11.002. Epub 2004 Dec 31. Exp Parasitol. 2005. PMID: 15639141
-
Molecular characteristics and physiology of microsporidia.Microbes Infect. 2001 Apr;3(5):407-15. doi: 10.1016/s1286-4579(01)01398-3. Microbes Infect. 2001. PMID: 11369278 Review.
-
Host-parasite coevolution: Insights from the Daphnia-parasite model system.Curr Opin Microbiol. 2008 Jun;11(3):290-301. doi: 10.1016/j.mib.2008.05.012. Epub 2008 Jun 13. Curr Opin Microbiol. 2008. PMID: 18556238 Review.
Cited by
-
Morphological and phylogenetic analysis of Nosema sp. HR (Microsporidia, Nosematidae): a new microsporidian pathogen of Histia rhodope Cramer (Lepidoptera, Zygaenidae).Parasitol Res. 2015 Mar;114(3):983-8. doi: 10.1007/s00436-014-4264-3. Epub 2014 Dec 25. Parasitol Res. 2015. PMID: 25538023
-
Near chromosome-level genome assembly of the microsporidium Hamiltosporidium tvaerminnensis.G3 (Bethesda). 2023 Sep 30;13(10):jkad185. doi: 10.1093/g3journal/jkad185. G3 (Bethesda). 2023. PMID: 37565496 Free PMC article.
-
Reduction and expansion in microsporidian genome evolution: new insights from comparative genomics.Genome Biol Evol. 2013;5(12):2285-303. doi: 10.1093/gbe/evt184. Genome Biol Evol. 2013. PMID: 24259309 Free PMC article.
-
Complete genome of a unicellular parasite (Antonospora locustae) and transcriptional interactions with its host locust.Microb Genom. 2020 Sep;6(9):mgen000421. doi: 10.1099/mgen.0.000421. Epub 2020 Aug 12. Microb Genom. 2020. PMID: 32783805 Free PMC article.
-
Contrasting host-pathogen interactions and genome evolution in two generalist and specialist microsporidian pathogens of mosquitoes.Nat Commun. 2015 May 13;6:7121. doi: 10.1038/ncomms8121. Nat Commun. 2015. PMID: 25968466 Free PMC article.
References
-
- Becnel JJ, Andreadis TG. In: The Microsporidia and Microsporidiosis. Witter M, Weiss LM, editor. Washington, DC: American Society of Microbiology Press; 1999. Microsporidia in insects. pp. 447–501.
-
- Larsson JIR. Identification of microsporidia. Acta Protozoologica. 1999;38:161–197.
-
- Vávra J, Larsson JIR. In: The Microsporidia and Microsporidiosis. Wittner M, Weiss LM, editor. Washington, DC: ASM Press; 1999. Structure of the microsporidia. pp. 7–84.
-
- Ishihara R, Hayashi YJ. Some properties of ribosomes from the sporoplasm of Nosema bombycis. Invert Pathol. 1968;11:377–385. doi: 10.1016/0022-2011(68)90186-9. - DOI
-
- Curgy JJ, Vávra J, Vivarès CP. Presence of ribosomal RNAs with prokaryotic properties in Microsporidia, eukaryotic organisms. Biol Cell. 1980;38:49–51.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

