miRNAs in lung cancer - studying complex fingerprints in patient's blood cells by microarray experiments
- PMID: 19807914
- PMCID: PMC2764731
- DOI: 10.1186/1471-2407-9-353
miRNAs in lung cancer - studying complex fingerprints in patient's blood cells by microarray experiments
Abstract
Background: Deregulated miRNAs are found in cancer cells and recently in blood cells of cancer patients. Due to their inherent stability miRNAs may offer themselves for blood based tumor diagnosis. Here we addressed the question whether there is a sufficient number of miRNAs deregulated in blood cells of cancer patients to be able to distinguish between cancer patients and controls.
Methods: We synthesized 866 human miRNAs and miRNA star sequences as annotated in the Sanger miRBase onto a microarray designed by febit biomed gmbh. Using the fully automated Geniom Real Time Analyzer platform, we analyzed the miRNA expression in 17 blood cell samples of patients with non-small cell lung carcinomas (NSCLC) and in 19 blood samples of healthy controls.
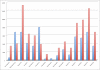
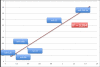
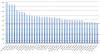
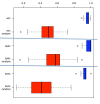
Results: Using t-test, we detected 27 miRNAs significantly deregulated in blood cells of lung cancer patients as compared to the controls. Some of these miRNAs were validated using qRT-PCR. To estimate the value of each deregulated miRNA, we grouped all miRNAs according to their diagnostic information that was measured by Mutual Information. Using a subset of 24 miRNAs, a radial basis function Support Vector Machine allowed for discriminating between blood cell samples of tumor patients and controls with an accuracy of 95.4% [94.9%-95.9%], a specificity of 98.1% [97.3%-98.8%], and a sensitivity of 92.5% [91.8%-92.5%].
Conclusion: Our findings support the idea that neoplasia may lead to a deregulation of miRNA expression in blood cells of cancer patients compared to blood cells of healthy individuals. Furthermore, we provide evidence that miRNA patterns can be used to detect human cancers from blood cells.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

