T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow
- PMID: 19808259
- PMCID: PMC2768857
- DOI: 10.1084/jem.20090525
T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow
Abstract
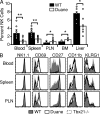
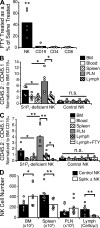
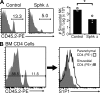
During a screen for ethylnitrosourea-induced mutations in mice affecting blood natural killer (NK) cells, we identified a strain, designated Duane, in which NK cells were reduced in blood and spleen but increased in lymph nodes (LNs) and bone marrow (BM). The accumulation of NK cells in LNs reflected a decreased ability to exit into lymph. This strain carries a point mutation within Tbx21 (T-bet), which generates a defective protein. Duane NK cells have a 30-fold deficiency in sphingosine-1-phosphate receptor 5 (S1P5) transcript levels, and S1P5-deficient mice exhibit an egress defect similar to Duane. Chromatin immunoprecipitation confirms binding of T-bet to the S1pr5 locus. S1P-deficient mice exhibit a more severe NK cell egress block, and the FTY720-sensitive S1P1 also plays a role in NK cell egress from LNs. S1P5 is not inhibited by CD69, a property that may facilitate trafficking of activated NK cells to effector sites. Finally, the accumulation of NK cells within BM of S1P-deficient mice was associated with reduced numbers in BM sinusoids, suggesting a role for S1P in BM egress. In summary, these findings identify S1P5 as a T-bet-induced gene that is required for NK cell egress from LNs and BM.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- AI52127/AI/NIAID NIH HHS/United States
- DA019674/DA/NIDA NIH HHS/United States
- BAA-NIH-NIAID-DAIT-07-35/PHS HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 AI052127/AI/NIAID NIH HHS/United States
- T3207270/PHS HHS/United States
- R01 AI042370/AI/NIAID NIH HHS/United States
- AI74847/AI/NIAID NIH HHS/United States
- R01 DA019674/DA/NIDA NIH HHS/United States
- R01 AI076458/AI/NIAID NIH HHS/United States
- R01 AI061061/AI/NIAID NIH HHS/United States
- R01 AI074847/AI/NIAID NIH HHS/United States
- R56 AI061061/AI/NIAID NIH HHS/United States
- AI061061/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

