Control of fetal hemoglobin: new insights emerging from genomics and clinical implications
- PMID: 19808799
- PMCID: PMC2758709
- DOI: 10.1093/hmg/ddp401
Control of fetal hemoglobin: new insights emerging from genomics and clinical implications
Abstract
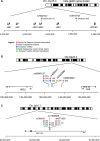
Increased levels of fetal hemoglobin (HbF, alpha(2)gamma(2)) are of no consequence in healthy adults, but confer major clinical benefits in patients with sickle cell anemia (SCA) and beta thalassemia, diseases that represent major public health problems. Inter-individual HbF variation is largely genetically controlled, with one extreme caused by mutations involving the beta globin gene (HBB) complex, historically referred to as pancellular hereditary persistence of fetal hemoglobin (HPFH). These Mendelian forms of HPFH are rare and do not explain the common form of heterocellular HPFH which represents the upper tail of normal HbF variation, and is clearly inherited as a quantitative genetic trait. Genetic studies have identified three major quantitative trait loci (QTLs) (Xmn1-HBG2, HBS1L-MYB intergenic region on chromosome 6q23, and BCL11A on chromosome 2p16) that account for 20-50% of the common variation in HbF levels in patients with SCA and beta thalassemia, and in healthy adults. Two of the major QTLs include oncogenes, emphasizing the importance of cell proliferation and differentiation as an important contribution to the HbF phenotype. The review traces the story of HbF quantitative genetics that uncannily mirrors the changing focus in genetic methodology, from candidate genes through positional cloning, to genome-wide association, that have expedited the dissection of the genetic architecture underlying HbF variability. These genetic results have already provided remarkable insights into molecular mechanisms that underlie the hemoglobin 'switch'.
Figures

References
-
- Stamatoyannopoulos G. Molecular and cellular basis of hemeoglobin switching. In: Steinberg M.H., Forget B.G., Higgs D.R., Nagel R.L., editors. Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. Cambridge, UK: Cambridge University Press; 2001. pp. 131–145.
-
- Wood W.G. Hereditary persistence of fetal hemoglobin and delta beta thalassemia. In: Steinberg M.H., Forget B.G., Higgs D.R., Nagel R.L., editors. Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. Cambridge, UK: Cambridge University Press; 2001. pp. 356–388.
-
- Weatherall D.J. Phenotype-genotype relationships in monogenic disease: lessons from the thalassaemias. Nat. Rev. Genet. 2001;2:245–255. - PubMed
-
- Stuart M.J., Nagel R.L. Sickle-cell disease. Lancet. 2004;364:1343–1360. - PubMed
-
- Thein S.L. Genetic modifiers of the beta-haemoglobinopathies. Br. J. Haematol. 2008;141:357–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

