A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice
- PMID: 19808965
- PMCID: PMC3700628
- DOI: 10.1158/0008-5472.CAN-09-1135
A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice
Abstract
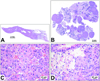
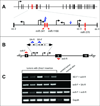
Recent advances in cancer therapeutics stress the need for a better understanding of the molecular mechanisms driving tumor formation. This can be accomplished by obtaining a more complete description of the genes that contribute to cancer. We previously described an approach using the Sleeping Beauty (SB) transposon system to model hematopoietic malignancies in mice. Here, we describe modifications of the SB system that provide additional flexibility in generating mouse models of cancer. First, we describe a Cre-inducible SBase allele, RosaSBase(LsL), that allows the restriction of transposon mutagenesis to a specific tissue of interest. This allele was used to generate a model of germinal center B-cell lymphoma by activating SBase expression with an Aid-Cre allele. In a second approach, a novel transposon was generated, T2/Onc3, in which the CMV enhancer/chicken beta-actin promoter drives oncogene expression. When combined with ubiquitous SBase expression, the T2/Onc3 transposon produced nearly 200 independent tumors of more than 20 different types in a cohort of 62 mice. Analysis of transposon insertion sites identified novel candidate genes, including Zmiz1 and Rian, involved in squamous cell carcinoma and hepatocellular carcinoma, respectively. These novel alleles provide additional tools for the SB system and provide some insight into how this mutagenesis system can be manipulated to model cancer in mice.
Figures


References
-
- Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science (New York, NY. 2007;318:1108–1113. - PubMed
-
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

