Limited copy number-high resolution melting (LCN-HRM) enables the detection and identification by sequencing of low level mutations in cancer biopsies
- PMID: 19811662
- PMCID: PMC2766370
- DOI: 10.1186/1476-4598-8-82
Limited copy number-high resolution melting (LCN-HRM) enables the detection and identification by sequencing of low level mutations in cancer biopsies
Abstract
Background: Mutation detection in clinical tumour samples is challenging when the proportion of tumour cells, and thus mutant alleles, is low. The limited sensitivity of conventional sequencing necessitates the adoption of more sensitive approaches. High resolution melting (HRM) is more sensitive than sequencing but identification of the mutation is desirable, particularly when it is important to discriminate false positives due to PCR errors or template degradation from true mutations.We thus developed limited copy number - high resolution melting (LCN-HRM) which applies limiting dilution to HRM. Multiple replicate reactions with a limited number of target sequences per reaction allow low level mutations to be detected. The dilutions used (based on Ct values) are chosen such that mutations, if present, can be detected by the direct sequencing of amplicons with aberrant melting patterns.
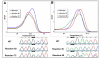
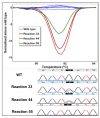
Results: Using cell lines heterozygous for mutations, we found that the mutations were not readily detected when they comprised 10% of total alleles (20% tumour cells) by sequencing, whereas they were readily detectable at 5% total alleles by standard HRM. LCN-HRM allowed these mutations to be identified by direct sequencing of those positive reactions.LCN-HRM was then used to review formalin-fixed paraffin-embedded (FFPE) clinical samples showing discordant findings between sequencing and HRM for KRAS exon 2 and EGFR exons 19 and 21. Both true mutations present at low levels and sequence changes due to artefacts were detected by LCN-HRM. The use of high fidelity polymerases showed that the majority of the artefacts were derived from the damaged template rather than replication errors during amplification.
Conclusion: LCN-HRM bridges the sensitivity gap between HRM and sequencing and is effective in distinguishing between artefacts and true mutations.
Figures


References
-
- Kobelt DJ, Hees SM, Hildebrandt RD, Wnendt S. Validation of a Method for Automated Nucleotide Sequence Analysis and Estimation of the Limits of Detection of Variant Sequences in a Heterogenic DNA Sample. Analytical Letters. 1998;31:41–54.
-
- Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli F, Mezzetti A, et al. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

