Structural and functional-annotation of an equine whole genome oligoarray
- PMID: 19811692
- PMCID: PMC3226197
- DOI: 10.1186/1471-2105-10-S11-S8
Structural and functional-annotation of an equine whole genome oligoarray
Abstract
Background: The horse genome is sequenced, allowing equine researchers to use high-throughput functional genomics platforms such as microarrays; next-generation sequencing for gene expression and proteomics. However, for researchers to derive value from these functional genomics datasets, they must be able to model this data in biologically relevant ways; to do so requires that the equine genome be more fully annotated. There are two interrelated types of genomic annotation: structural and functional. Structural annotation is delineating and demarcating the genomic elements (such as genes, promoters, and regulatory elements). Functional annotation is assigning function to structural elements. The Gene Ontology (GO) is the de facto standard for functional annotation, and is routinely used as a basis for modelling and hypothesis testing, large functional genomics datasets.
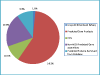
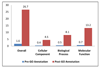
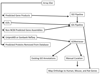
Results: An Equine Whole Genome Oligonucleotide (EWGO) array with 21,351 elements was developed at Texas A&M University. This 70-mer oligoarray was designed using the approximately 7 x assembled and annotated sequence of the equine genome to be one of the most comprehensive arrays available for expressed equine sequences. To assist researchers in determining the biological meaning of data derived from this array, we have structurally annotated it by mapping the elements to multiple database accessions, including UniProtKB, Entrez Gene, NRPD (Non-Redundant Protein Database) and UniGene. We next provided GO functional annotations for the gene transcripts represented on this array. Overall, we GO annotated 14,531 gene products (68.1% of the gene products represented on the EWGO array) with 57,912 annotations. GAQ (GO Annotation Quality) scores were calculated for this array both before and after we added GO annotation. The additional annotations improved the meanGAQ score 16-fold. This data is publicly available at AgBase http://www.agbase.msstate.edu/.
Conclusion: Providing additional information about the public databases which link to the gene products represented on the array allows users more flexibility when using gene expression modelling and hypothesis-testing computational tools. Moreover, since different databases provide different types of information, users have access to multiple data sources. In addition, our GO annotation underpins functional modelling for most gene expression analysis tools and enables equine researchers to model large lists of differentially expressed transcripts in biologically relevant ways.
Figures

References
-
- Horse Genome Assembled. http://www.genome.gov/20519480
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

