The structural changes of T7 RNA polymerase from transcription initiation to elongation
- PMID: 19811903
- PMCID: PMC2818687
- DOI: 10.1016/j.sbi.2009.09.001
The structural changes of T7 RNA polymerase from transcription initiation to elongation
Abstract
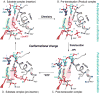
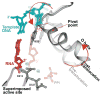
The structures of T7 RNA polymerase (T7 RNAP) captured in the initiation and elongation phases of transcription, as well as an intermediate stage provide insights into how this RNA polymerase protein can initiate RNA synthesis and synthesize 7-10 nucleotides of RNA while remaining bound to the DNA promoter site. Recently, the structures of T7 RNAP bound to its promoter DNA along with either a seven nucleotide or eight nucleotide transcript show an elongated product site resulting from a 40 degrees or 45 degrees rotation of the promoter and domain that binds it. The different functional properties of the initiation and elongation phases of transcription are illuminated from structures of the initiation and elongation complexes. Structural insights into the translocation of the product transcript of RNAP, its separation of the downstream duplex DNA, and its removal of the transcript from the heteroduplex are provided by the structures of several states of nucleotide incorporation. A conformational change in the 'fingers' domain that results from the binding or dissociation of incoming NTP or PPi appears to be associated with the state of translocation of T7 RNAP.
Figures

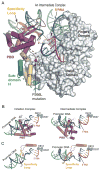
References
-
- Cheetham GMT, Steitz TA. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305. - PubMed
-
- Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387. - PubMed
-
- Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyoma S. Structure of a T7 RNA polymerase elongation complex at 2.9 Å resolution. Nature. 2002;420:43. - PubMed
-
- Durniak K, Bailey S, Steitz TA. The structure of a transcribing T7 RNA polymerase in transition from initiation to elongation. Science. 2008;322:553–557. The crystal structures of T7 RNAP bound to substrate DNA and either a 7 nt or 8nt transcript are described. They show a 40° and 45° rotation of the promoter binding domains resulting in an enlargement of the heteroduplex product binding cleft. They represent intermediate states in the transition from the initiation to elongation states. - PMC - PubMed
-
- Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

