Identification of hepatitis C virus NS5A inhibitors
- PMID: 19812153
- PMCID: PMC2798423
- DOI: 10.1128/JVI.01360-09
Identification of hepatitis C virus NS5A inhibitors
Abstract
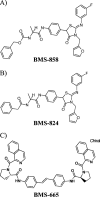
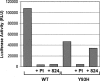
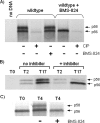
Using a cell-based replicon screen, we identified a class of compounds with a thiazolidinone core structure as inhibitors of hepatitis C virus (HCV) replication. The concentration of one such compound, BMS-824, that resulted in a 50% inhibition of HCV replicon replication was approximately 5 nM, with a therapeutic index of >10,000. The compound showed good specificity for HCV, as it was not active against several other RNA and DNA viruses. Replicon cells resistant to BMS-824 were isolated, and mutations were identified. A combination of amino acid substitutions of leucine to valine at residue 31 (L31V) and glutamine to leucine at residue 54 (Q54L) in NS5A conferred resistance to this chemotype, as did a single substitution of tyrosine to histidine at amino acid 93 (Y93H) in NS5A. To further explore the region(s) of NS5A involved in inhibitor sensitivity, genotype-specific NS5A inhibitors were used to evaluate a series of genotype 1a/1b hybrid replicons. Our results showed that, consistent with resistance mapping, the inhibitor sensitivity domain also mapped to the N terminus of NS5A, but it could be distinguished from the key resistance sites. In addition, we demonstrated that NS5A inhibitors, as well as an active-site inhibitor that specifically binds NS3 protease, could block the hyperphosphorylation of NS5A, which is believed to play an essential role in the viral life cycle. Clinical proof of concept has recently been achieved with derivatives of these NS5A inhibitors, indicating that small molecules targeting a nontraditional viral protein like NS5A, without any known enzymatic activity, can also have profound antiviral effects on HCV-infected subjects.
Figures



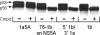
References
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Cheng, G., A. Montero, P. Gastaminza, C. Whitten-Bauer, S. F. Wieland, M. Isogawa, B. Fredericksen, S. Selvarajah, P. A. Gallay, M. R. Ghadiri, and F. V. Chisari. 2008. A virocidal amphipathic α-helical peptide that inhibits hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 105:3088-3093. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

