Characterization of viral and human RNAs smaller than canonical MicroRNAs
- PMID: 19812168
- PMCID: PMC2786840
- DOI: 10.1128/JVI.01325-09
Characterization of viral and human RNAs smaller than canonical MicroRNAs
Abstract
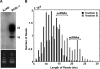
Recently identified small (20 to 40 bases) RNAs, such as microRNAs (miRNAs) and endogenous small interfering RNAs (siRNAs) participate in important cellular pathways. In this report, we systematically characterized several novel features of human and viral RNA products smaller than miRNAs. We found that Kaposi sarcoma-associated herpesvirus K12-1 miRNA (23 bases) associates with a distinct, unusually small (17-base) RNA (usRNA) that can effectively downregulate a K12-1 miRNA target, human RAD21, suggesting that stable degradation-like products may also contribute to gene regulation. High-throughput sequencing reveals a diverse set of human miRNA-derived usRNAs and other non-miRNA-derived usRNAs. Human miRNA-derived usRNAs preferentially match to 5' ends of miRNAs and are also more likely to associate with the siRNA effector protein Ago2 than with Ago1. Many non-miRNA-derived usRNAs associate with Ago proteins and also frequently contain C-rich 3'-specific motifs that are overrepresented in comparison to Piwi-interacting RNAs and transcription start site-associated RNAs. We postulate that approximately 30% of usRNAs could have evolved to participate in biological processes, including gene silencing.
Figures




References
-
- Reference deleted.
-
- Beitzinger, M., L. Peters, J. Y. Zhu, E. Kremmer, and G. Meister. 2007. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 4:76-84. - PubMed
-
- Berezikov, E., G. van Tetering, M. Verheul, J. van de Belt, L. van Laake, J. Vos, R. Verloop, M. van de Wetering, V. Guryev, S. Takada, A. J. van Zonneveld, H. Mano, R. Plasterk, and E. Cuppen. 2006. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 16:1289-1298. - PMC - PubMed
-
- Brennecke, J., A. A. Aravin, A. Stark, M. Dus, M. Kellis, R. Sachidanandam, and G. J. Hannon. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089-1103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

