Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription
- PMID: 19812244
- PMCID: PMC2785733
- DOI: 10.1091/mbc.e09-05-0380
Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription
Abstract
On entry into mitosis, many transcription factors dissociate from chromatin, resulting in global transcriptional shutdown. During mitosis, some genes are marked to ensure the inheritance of their expression in the next generation of cells. The nature of mitotic gene marking, however, has been obscure. Brd4 is a double bromodomain protein that localizes to chromosomes during mitosis and is implicated in holding mitotic memory. In interphase, Brd4 interacts with P-TEFb and functions as a global transcriptional coactivator. We found that throughout mitosis, Brd4 remained bound to the transcription start sites of many M/G1 genes that are programmed to be expressed at the end of, or immediately after mitosis. In contrast, Brd4 did not bind to genes that are expressed at later phases of cell cycle. Brd4 binding to M/G1 genes increased at telophase, the end phase of mitosis, coinciding with increased acetylation of histone H3 and H4 in these genes. Increased Brd4 binding was accompanied by the recruitment of P-TEFb and de novo M/G1 gene transcription, the events impaired in Brd4 knockdown cells. In sum, Brd4 marks M/G1 genes for transcriptional memory during mitosis, and upon exiting mitosis, this mark acts as a signal for initiating their prompt transcription in daughter cells.
Figures
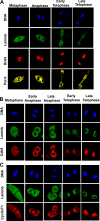
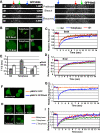
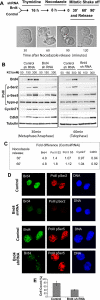

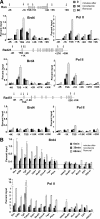

References
-
- Christova R., Oelgeschlager T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 2002;4:79–82. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

