Text mining and manual curation of chemical-gene-disease networks for the comparative toxicogenomics database (CTD)
- PMID: 19814812
- PMCID: PMC2768719
- DOI: 10.1186/1471-2105-10-326
Text mining and manual curation of chemical-gene-disease networks for the comparative toxicogenomics database (CTD)
Abstract
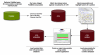
Background: The Comparative Toxicogenomics Database (CTD) is a publicly available resource that promotes understanding about the etiology of environmental diseases. It provides manually curated chemical-gene/protein interactions and chemical- and gene-disease relationships from the peer-reviewed, published literature. The goals of the research reported here were to establish a baseline analysis of current CTD curation, develop a text-mining prototype from readily available open source components, and evaluate its potential value in augmenting curation efficiency and increasing data coverage.
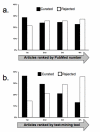
Results: Prototype text-mining applications were developed and evaluated using a CTD data set consisting of manually curated molecular interactions and relationships from 1,600 documents. Preliminary results indicated that the prototype found 80% of the gene, chemical, and disease terms appearing in curated interactions. These terms were used to re-rank documents for curation, resulting in increases in mean average precision (63% for the baseline vs. 73% for a rule-based re-ranking), and in the correlation coefficient of rank vs. number of curatable interactions per document (baseline 0.14 vs. 0.38 for the rule-based re-ranking).
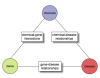
Conclusion: This text-mining project is unique in its integration of existing tools into a single workflow with direct application to CTD. We performed a baseline assessment of the inter-curator consistency and coverage in CTD, which allowed us to measure the potential of these integrated tools to improve prioritization of journal articles for manual curation. Our study presents a feasible and cost-effective approach for developing a text mining solution to enhance manual curation throughput and efficiency.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

